Abstract
Objective
Orofacial development is a complex process subjected to failure impairing. Indeed, the cleft of the lip and/or of the palate is among the most frequent inborn malformations. The JARID2 gene has been suggested to be involved in non-syndromic cleft lip with or without cleft palate (nsCL/P) etiology. JARID2 interacts with the polycomb repressive complex 2 (PRC2) in regulating the expression patterns of developmental genes by modifying the chromatin state.
Materials and methods
Genes coding for the PRC2 components, as well as other genes active in cell differentiation and embryonic development, were selected for a family-based association study to verify their involvement in nsCL/P. A total of 632 families from Italy and Asia participated to the study.
Results
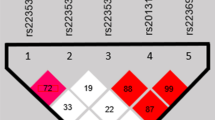
Evidence of allelic association was found with polymorphisms of SNAI1; in particular, the rs16995010-G allele was undertransmitted to the nsCL/P cases [P = 0.004, odds ratio = 0.69 (95% C.I. 0.54–0.89)]. However, the adjusted significance value corrected for all the performed tests was P = 0.051.
Conclusions
The findings emerging by the present study suggest for the first time an involvement of SNAI1 in the nsCL/P onset.
Clinical relevance
Interestingly, SNAI1 is known to promote epithelial to mesenchymal transition by repressing E-cadherin expression, but it needs an intact PRC2 to act this function. Alterations of this process could contribute to the complex etiology of nsCL/P.
Similar content being viewed by others
References
Calzolari E, Pierini A, Astolfi G, Bianchi F, Neville AJ, Rivieri F (2007) Associated anomalies in multi-malformed infants with cleft lip and palate: an epidemiologic study of nearly 6 million births in 23 EUROCAT registries. Am J Med Genet A 143A(6):528–537. https://doi.org/10.1002/ajmg.a.31447
Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC (2009) Cleft lip and palate. Lancet 374(9703):1773–1785. https://doi.org/10.1016/S0140-6736(09)60695-4
Lidral AC, Moreno LM (2005) Progress toward discerning the genetics of cleft lip. Curr Opin Pediatr 17(6):731–739. https://doi.org/10.1097/01.mop.0000185138.65820.7f
Beaty TH, Marazita ML, Leslie EJ (2016) Genetic factors influencing risk to orofacial clefts: today's challenges and tomorrow’s opportunities. F1000Res 5:2800. https://doi.org/10.12688/f1000research.9503.1
Eiberg H, Bixler D, Nielsen LS, Conneally PM, Mohr J (1987) Suggestion of linkage of a major locus for nonsyndromic orofacial cleft with F13A and tentative assignment to chromosome 6. Clin Genet 32(2):129–132
Scapoli L, Pezzetti F, Carinci F, Martinelli M, Carinci P, Tognon M (1997) Evidence of linkage to 6p23 and genetic heterogeneity in nonsyndromic cleft lip with or without cleft palate. Genomics 43(2):216–220. https://doi.org/10.1006/geno.1997.4798
Scapoli L, Martinelli M, Pezzetti F, Palmieri A, Girardi A, Savoia A, Bianco AM, Carinci F (2010) Expression and association data strongly support JARID2 involvement in nonsyndromic cleft lip with or without cleft palate. Hum Mutat 31(7):794–800. https://doi.org/10.1002/humu.21266
Hao Y, Mi N, Jiao X, Zheng X, Song T, Zhuang D, Tian S, Feng D (2015) Association of JARID2 polymorphisms with non-syndromic orofacial clefts in northern Chinese Han population. J Oral Pathol Med 44(5):386–391. https://doi.org/10.1111/jop.12244
Messetti AC, Machado RA, de Oliveira CE, Martelli-Junior H, de Almeida Reis SR, Moreira HS, Persuhn DC, Wu T, Coletta RD (2017) Brazilian multicenter study of association between polymorphisms in CRISPLD2 and JARID2 and non-syndromic oral clefts. J Oral Pathol Med 46(3):232–239. https://doi.org/10.1111/jop.12470
Martinez AM, Cavalli G (2006) The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle 5(11):1189–1197. https://doi.org/10.4161/cc.5.11.2781
Surface LE, Thornton SR, Boyer LA (2010) Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7(3):288–298. https://doi.org/10.1016/j.stem.2010.08.004
Prezioso C, Orlando V (2011) Polycomb proteins in mammalian cell differentiation and plasticity. FEBS Lett 585(13):2067–2077. https://doi.org/10.1016/j.febslet.2011.04.062
Piunti A, Pasini D (2011) Epigenetic factors in cancer development: polycomb group proteins. Future Oncol 7(1):57–75. https://doi.org/10.2217/fon.10.157
Pontier DB, Gribnau J (2011) Xist regulation and function explored. Hum Genet 130(2):223–236. https://doi.org/10.1007/s00439-011-1008-7
O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21(13):4330–4336. https://doi.org/10.1128/MCB.21.13.4330-4336.2001
Schuettengruber B, Cavalli G (2009) Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136(21):3531–3542. https://doi.org/10.1242/dev.033902
Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10(10):697–708. https://doi.org/10.1038/nrm2763
Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464(7286):306–310. https://doi.org/10.1038/nature08788
Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139(7):1303–1314. https://doi.org/10.1016/j.cell.2009.12.003
Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24(4):368–380. https://doi.org/10.1101/gad.1886410
Cooper S, Grijzenhout A, Underwood E, Ancelin K, Zhang T, Nesterova TB, Anil-Kirmizitas B, Bassett A, Kooistra SM, Agger K, Helin K, Heard E, Brockdorff N (2016) Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun 7:13661. https://doi.org/10.1038/ncomms13661
Lin Y, Dong C, Zhou BP (2014) Epigenetic regulation of EMT: the Snail story. Curr Pharm Des 20(11):1698–1705. https://doi.org/10.2174/13816128113199990512
Tange S, Oktyabri D, Terashima M, Ishimura A, Suzuki T (2014) JARID2 is involved in transforming growth factor-beta-induced epithelial-mesenchymal transition of lung and colon cancer cell lines. PLoS One 9(12):e115684. https://doi.org/10.1371/journal.pone.0115684
Nouri N, Memarzadeh M, Carinci F, Cura F, Scapoli L, Nouri N, Jafary F, Sedghi M, Sadri L, Salehi M (2015) Family-based association analysis between nonsyndromic cleft lip with or without cleft palate and IRF6 polymorphism in an Iranian population. Clin Oral Investig 19(4):891–894. https://doi.org/10.1007/s00784-014-1305-3
Martinelli M, Girardi A, Cura F, Nouri N, Pinto V, Carinci F, Morselli PG, Salehi M, Scapoli L (2016) Non-syndromic cleft lip with or without cleft palate in Asian populations: association analysis on three gene polymorphisms of the folate pathway. Arch Oral Biol 61:79–82. https://doi.org/10.1016/j.archoralbio.2015.10.019
Koivisto AM, Ala-Mello S, Lemmela S, Komu HA, Rautio J, Jarvela I (2007) Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin Genet 72(2):145–149. https://doi.org/10.1111/j.1399-0004.2007.00836.x
Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, Garcia de Herreros A, Peiro S (2008) Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 28(15):4772–4781. https://doi.org/10.1128/MCB.00323-08
de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D (2005) Efficiency and power in genetic association studies. Nat Genet 37(11):1217–1223. https://doi.org/10.1038/ng1669
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. https://doi.org/10.1093/bioinformatics/bth457
Cura F, Bohmer AC, Klamt J, Schunke H, Scapoli L, Martinelli M, Carinci F, Nothen MM, Knapp M, Ludwig KU, Mangold E (2016) Replication analysis of 15 susceptibility loci for nonsyndromic cleft lip with or without cleft palate in an italian population. Birth Defects Res A Clin Mol Teratol 106(2):81–87. https://doi.org/10.1002/bdra.23454
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575. https://doi.org/10.1086/519795
Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52(3):506–516
Kazeem GR, Farrall M (2005) Integrating case-control and TDT studies. Ann Hum Genet 69(3):329–335. https://doi.org/10.1046/J.1469-1809.2005.00156.x
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:289–300
Mehrotra D (2015) Genomic expression in non syndromic cleft lip and palate patients: a review. J Oral Biol Craniofac Res 5(2):86–91. https://doi.org/10.1016/j.jobcr.2015.03.003
Carver EA, Jiang R, Lan Y, Oram KF, Gridley T (2001) The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol 21(23):8184–8188. https://doi.org/10.1128/MCB.21.23.8184-8188.2001
Noden DM (1986) Origins and patterning of craniofacial mesenchymal tissues. J Craniofac Genet Dev Biol Suppl 2:15–31
Hay ED (2005) The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn 233(3):706–720. https://doi.org/10.1002/dvdy.20345
Yu W, Ruest LB, Svoboda KK (2009) Regulation of epithelial-mesenchymal transition in palatal fusion. Exp Biol Med (Maywood) 234(5):483–491. https://doi.org/10.3181/0812-MR-365
Martinez-Alvarez C, Blanco MJ, Perez R, Rabadan MA, Aparicio M, Resel E, Martinez T, Nieto MA (2004) Snail family members and cell survival in physiological and pathological cleft palates. Dev Biol 265(1):207–218. https://doi.org/10.1016/j.ydbio.2003.09.022
Yu W, Zhang Y, Ruest LB, Svoboda KK (2013) Analysis of Snail1 function and regulation by Twist1 in palatal fusion. Front Physiol 4:12
Murray SA, Oram KF, Gridley T (2007) Multiple functions of Snail family genes during palate development in mice. Development 134(9):1789–1797. https://doi.org/10.1242/dev.02837
Miller SF, Weinberg SM, Nidey NL, Defay DK, Marazita ML, Wehby GL, Moreno Uribe LM (2014) Exploratory genotype-phenotype correlations of facial form and asymmetry in unaffected relatives of children with non-syndromic cleft lip and/or palate. J Anat 224(6):688–709. https://doi.org/10.1111/joa.12182
Song H, Wang X, Yan J, Mi N, Jiao X, Hao Y, Zhang W, Gao Y (2017) Association of single-nucleotide polymorphisms of CDH1 with nonsyndromic cleft lip with or without cleft palate in a northern Chinese Han population. Medicine (Baltimore) 96(5):e5574. https://doi.org/10.1097/MD.0000000000005574
Martinelli M, Carinci F, Morselli PG, Caramelli E, Palmieri A, Girardi A, Riberti C, Scapoli L (2011) Evidence of LEF1 fetal-maternal interaction in cleft lip with or without cleft palate in a consistent Italian sample study. Int J Immunopathol Pharmacol 24(2_suppl):15–19. https://doi.org/10.1177/03946320110240S204
Acknowledgements
We are indebted to the families participating in the study for their invaluable contribution, as well as to all the personnel involved in clinical data and specimen collection.
Funding
This work was supported in part by a grant from the Association Interethnos Interplast Italy and by Fondazione Del Monte di Bologna e Ravenna (for the salary of Dr. Ambra Girardi).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
This investigation collected evidence supporting a possible effect of SNAI1 polymorphisms in nsCL/P etiology.
Luca Scapoli and Marcella Martinelli have shared senior authorship.
Rights and permissions
About this article
Cite this article
Cura, F., Palmieri, A., Girardi, A. et al. Possible effect of SNAIL family transcriptional repressor 1 polymorphisms in non-syndromic cleft lip with or without cleft palate. Clin Oral Invest 22, 2535–2541 (2018). https://doi.org/10.1007/s00784-018-2350-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-018-2350-0