Abstract
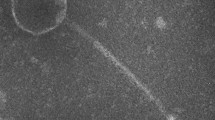
Phage Vp_R1 belongs to the family Podoviridae and has a C3 morphotype, with an elongated head with a diameter of 190 ± 1.1 nm and an ultrashort tail with a length of 9 ± 1.2 nm. The double-stranded DNA genome is 112.1 kb long, has a mol% G + C content of 40.3, contains 129 ORFs, and encodes four tRNAs. Phylogenetic analysis suggests that phage Vp_R1 is a novel member of the genus Kuravirus.




Similar content being viewed by others
References
Ackermann HW (2001) Frequency of morphological phage descriptions in the year 2000. Brief review. Arch Virol 146:843–857
Ackermann HW (2007) 5500 Phages examined in the electron microscope. Arch Virol 152:227–243
Ceccarelli D, Hasan NA, Huq A, Colwell RR (2013) Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front Cell Infect Mi 3:97
Chen D, Texada DE (2006) Low-usage codons and rare codons of Escherichia coli mini review. Gene Ther Mol Biol 10:1–12
Chen Y, Liu XM, Yan JW, Li XG, Mei LL, Mao QF, Ma Y (2010) Foodborne pathogens in retail oysters in south China. Biomed Environ Sci 23:32–36
Chopin A, Deveau H, Ehrlich SD, Moineau S, Chopin M (2007) KSY1, a lactococcal phage with a T7-like transcription. Virology 365:1–9
Doss J, Culbertson K, Hahn D, Camacho J, Barekzi N (2017) A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses 9:50
Dwivedi B, Xue B, Lundin D, Edwards RA, Breitbart M (2013) A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biol 13:33
Goldsmith DB, Crosti G, Dwivedi B, McDaniel LD, Varsani A, Suttle CA, Weinbauer MG, Sandaa R, Breitbart M (2011) Development of phoH as a novel signature gene for assessing marine phage diversity. Appl Environ Microb 77:7730–7739
Goodridge L, Gallaccio A, Griffiths MW (2003) Morphological, host range, and genetic characterization of two coliphages. Appl Environ Microb 69:5364–5371
Harakudo Y, Saito S, Ohtsuka K, Yamasaki S, Yahiro S, Nishio T, Iwade Y, Otomo Y, Konuma H, Tanaka H (2012) Characteristics of a sharp decrease in vibrio parahaemolyticus infections and seafood contamination in Japan. Int J Food Microbiol 157:95–101
Kenshi M, Jumpei U, Shin-Ichiro K, Takako U, Hiroshi H, Shigeyoshi S, Asako M, Hiroshi W, Shigenobu M (2010) Morphological and genetic analysis of three bacteriophages of Serratia marcescens isolated from environmental water. FEMS Microbiol Lett 291:201–208
Kropinski AM, Lingohr EJ, Ackermann H (2011) The genome sequence of enterobacterial phage 7–11, which possesses an unusually elongated head. Arch Virol 156:149–151
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Lee JK, Jung DW, Eom SY, Oh SW, Kim Y, Kwak HS, Kim YH (2008) Occurrence of Vibrio parahaemolyticus in oysters from Korean retail outlets. Food Control 19:990–994
Lenneman B, Rothman-Denes L (2015) Structural and biochemical investigation of bacteriophage N4-encoded RNA polymerases. Biomolecules 5:647–667
Li Y, Chen M, Tang F, Yao H, Lu C, Zhang W (2012) Complete genome sequence of the novel lytic avian pathogenic coliphage NJ01. J Virol 86:13874–13875
Mirzaei MK, Eriksson H, Kasuga K, Haggård-Ljungquist E, Nilsson AS (2014) Genomic, proteomic, morphological, and phylogenetic analyses of vB_EcoP_SU10, a podoviridae phage with C3 morphology. PloS One 9:e116294
Nakai T, Park SC (2002) Bacteriophage therapy of infectious diseases in aquaculture. Res Microbiol 153:13–18
Ren H, Li Z, Xu Y, Wang L, Li X (2019) Protective effectiveness of feeding phage cocktails in controlling Vibrio parahaemolyticus infection of sea cucumber Apostichopus japonicus. Aquaculture 503:322–329
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, volume 2. Anal Biochem 186:182–183
Savalia D, Westblade LF, Goel M, Florens L, Kemp P, Akulenko N, Pavlova O, Padovan JC, Chait BT, Washburn MP, Ackermann H, Mushegian A, Gabisonia T, Molineux I, Severinov K (2008) Genomic and proteomic analysis of phiEco32, a novel Escherichia coli bacteriophage. J Mol Biol 377:774–789
Saxelin M, Nurmiaho E, Korhola MP, Sundman V (1979) Partial characterization of a new C3-type capsule-dissolving phage ofStreptococcus cremoris. Can J Microbiol 25:1182–1187
Zhang L, Orth K (2013) Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol 16:70–77
Acknowledgements
The authors would like to thank the members of the group of Professor Yigang Tong, Beijing Institute of Microbiology and Epidemiology, for help with sequence analysis.
Funding
This work was financially supported by the National Public Science and Technology Research Funds Projects of Ocean (Grant No. 201405003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals by any author.
Additional information
Handling Editor: T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1 Secondary structure visualization of tRNAs of Vp_R1. A. tRNA-Met (CAT). B. tRNA–Ile (GAT). C. tRNA- Arg (CCT). D. tRNA-Arg (TCT).
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ren, H., Li, Z., Xu, L. et al. Genome sequence analysis of Vibrio parahaemolyticus lytic phage Vp_R1 with a C3 morphotype. Arch Virol 164, 2865–2871 (2019). https://doi.org/10.1007/s00705-019-04364-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04364-0