Abstract
We aimed to enhance understanding of the molecular diversity of arbuscular mycorrhizal fungi (AMF) by building a new global dataset targeting previously unstudied geographical areas. In total, we sampled 96 plant species from 25 sites that encompassed all continents except Antarctica. AMF in plant roots were detected by sequencing the nuclear SSU rRNA gene fragment using either cloning followed by Sanger sequencing or 454-sequencing. A total of 204 AMF phylogroups (virtual taxa, VT) were recorded, increasing the described number of Glomeromycota VT from 308 to 341 globally. Novel VT were detected from 21 sites; three novel but nevertheless widespread VT (Glomus spp. MO-G52, MO-G53, MO-G57) were recorded from six continents. The largest increases in regional VT number were recorded in previously little-studied Oceania and in the boreal and polar climatic zones — this study providing the first molecular data from the latter. Ordination revealed differences in AM fungal communities between different continents and climatic zones, suggesting that both biogeographic history and environmental conditions underlie the global variation of those communities. Our results show that a considerable proportion of Glomeromycota diversity has been recorded in many regions, though further large increases in richness can be expected in remaining unstudied areas.



Similar content being viewed by others
Notes
The family-level taxonomy of Glomeromycota follows Schüßler and Walker (2010), unless otherwise stated.
New VT acquire numerical codes of MaarjAM VT nomenclature after data from a publication have been uploaded to the MaarjAM database.
References
Bago B, Pfeffer PE, Shachar-Hill Y (2000) Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol 124:949–957
Bik HM, Porazinska DL, Creer S, Caporaso JG, Knight R, Thomas WK (2012) Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol Evol 27:233–243
Brock PM, Döring H, Bidartondo MI (2009) How to know unknown fungi: the role of a herbarium. New Phytol 181:719–724
Chaudhary VB, Lau MK, Johnson NC (2008) Macroecology of microbes — biogeography of the Glomeromycota. In: Varma A (ed) Mycorrhiza: genetics and molecular biology, eco-function, biotechnology, eco-physiology, structure and systematics. Springer-Verlag, Berlin, pp 529–562
Clapp JP, Young JPW, Merryweather JW, Fitter AH (1995) Diversity of fungal symbionts in arbuscular mycorrhizas from a natural community. New Phytol 130:259–265
Davison J, Öpik M, Zobel M, Vasar M, Metsis M, Moora M (2012) Communities of arbuscular mycorrhizal fungi detecting in forest soil are spatially heterogeneous but do not vary throughout the growing season. PLoS One 7:e41938
de Souza FA, Kowalchuk GA, Leeflang P, van Veen JA, Smit E (2004) PCR-denaturing gradient gel electrophoresis profiling of inter- and intraspecies 18S rRNA gene sequence heterogeneity is an accurate and sensitive method to assess species diversity of arbuscular mycorrhizal fungi of the genus Gigaspora. Appl Environ Microbiol 70:1413–1424
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH (2010) Idiosyncrasy and overdominance in the structure of natural communities of arbuscular mycorrhizal fungi: is there a role for stochastic processes? J Ecol 98:419–428
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Fitter AH (2005) Darkness visible: reflections on underground ecology. J Ecol 93:231–243
Gamper HA, Walker C, Schüßler A (2009) Diversispora celata sp. nov: molecular ecology and phylotaxonomy of an inconspicuous arbuscular mycorrhizal fungus. New Phytol 182:495–506
Goto BT, Silva GA, de Assis DMA, Silva DKA, Souza RG, Ferreira ACA, Jobim K, Mello CMA, Vieira HEE, Maia LC, Oehl F (2012) Intraornatosporaceae (Gigasporales), a new family with two new genera and two new species. Mycotaxon 119:117–132
Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW (1998) Ploughing up the wood-wide web? Nature 394:431
Hibbett D, Glotzer D (2011) Where are all the undocumented fungal species? A study of Mortierella demonstrates the need for sequence-based classification. New Phytol 191:592–596
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH (2011) Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biol Rev 25:38–47
Hibbett DS, Ohman A, Kirk PM (2009) Fungal ecology catches fire. New Phytol 184:279–282
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298
Kivlin SN, Hawkes CV, Treseder KK (2011) Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biol Biochem 43:2294–2303
Krüger M, Krüger C, Walker C, Stockinger H, Schüßler A (2012) Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol 193:970–984
Krüger M, Stockinger H, Krüger C, Schüßler A (2009) DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol 183:212–223
Lee J, Lee S, Young JPW (2008) Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol Ecol 65:339–349
Lentendu G, Zinger L, Manel S, Coissac E, Choler P, Geremia RA, Melodelima C (2011) Assessment of soil fungal diversity in different alpine tundra habitats by means of pyrosequencing. Fungal Divers 49:113–123
Liu Y, He J, Shi G, An LZ, Öpik M, Feng HY (2011) Diverse communities of arbuscular mycorrhizal fungi inhabit sites with very high altitude in Tibet Plateau. FEMS Microbiol Ecol 78:355–365
Merckx VSFT, Janssens SB, Hynson NA, Specht CD, Bruns TD, Smets EF (2012) Mycoheterotrophic interactions are not limited to a narrow phylogenetic range of arbuscular mycorrhizal fungi. Mol Ecol 21:1524–1532
Merryweather J, Fitter A (1998) The arbuscular mycorrhizal fungi of Hyacinthoides non-scripta: I. Diversity of fungal taxa. New Phytol 138:117–129
Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshal DF, Wright F (2009) TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25:126–127
Moora M, Berger S, Davison J, Öpik M, Bommarco R, Bruelheide H, Kühn I, Kunin WE, Metsis M, Rortais A, Vanatoa A, Vanatoa E, Stout JC, Truusa M, Westphal C, Zobel M, Walther GR (2011) Alien plants associate with widespread generalist arbuscular mycorrhizal fungal taxa: evidence from a continental-scale study using massively parallel 454-sequencing. J Biogeogr 38:1305–1317
Morton JB, Bentivenga SP, Bever JD (1995) Discovery, measurement, and interpretation of diversity in arbuscular endomycorrhizal fungi (Glomales, Zygomycetes). Can J Bot 73:S25–S32
Nagy LG, Petkovits T, Kovács GM, Voigt K, Vágvölgyi C, Papp T (2011) Where is the unseen fungal diversity hidden? A study of Mortierella reveals a large contribution of reference collections to the identification of fungal environmental sequences. New Phytol 191:789–794
Newsham KK, Upson R, Read DJ (2009) Mycorrhizas and dark septate root endophytes in polar regions. Fungal Ecol 2:10–20
Oehl F, Da Silva GA, Goto BT, Sieverding E (2011a) Glomeromycota: three new genera and glomoid species reorganized. Mycotaxon 116:75–120
Oehl F, Da Silva GA, Sanchez-Castro I, Goto BT, Maia LC, Vieira HEE, Sieverding E, Palenzuela J (2011b) Revision of Glomeromycetes with entrophosporoid and glomoid spore formation with three new genera. Mycotaxon 117:297–316
Oehl F, Sieverding E, Palenzuela J, Ineichen K, da Silva GA (2011c) Advances in Glomeromycota taxonomy and classification. IMA Fungus 2:191–199
Oksanen J, Blanchet FG, Kindt R, Legendre P, O’Hara RG, Minchin PR, Simpson GL, Solymos PS, Henry MH, Wagner H (2011) Vegan: community ecology package, ver.2.02 http://cran.r-project.org http://vegan.r-forge.r-project.org/
Öpik M, Metsis M, Daniell TJ, Zobel M, Moora M (2009) Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol 184:424–437
Öpik M, Moora M, Liira J, Kõljalg U, Zobel M, Sen R (2003) Divergent arbuscular mycorrhizal fungal communities colonize roots of Pulsatilla spp. in boreal Scots pine forest and grassland soils. New Phytol 160:581–593
Öpik M, Moora M, Liira J, Zobel M (2006) Composition of root-colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J Ecol 94:778–790
Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier Ü, Zobel M (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188:223–241
Parameswaran P, Jalili R, Tao L, Shokralla S, Harizadeh B, Ronaghi M, Fire AZ (2007) A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res 35:e130
Peay KG, Bidartondo MI, Arnold AE (2010) Not every fungus is everywhere: scaling to the biogeography of fungal–plant interactions across roots, shoots and ecosystems. New Phytol 185:878–882
Pietikäinen A, Kytöviita MM, Husband R, Young JPW (2007) Diversity and persistence of arbuscular mycorrhizas in a low-Arctic meadow habitat. New Phytol 176:691–698
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Powell JR, Monaghan MT, Öpik M, Rillig MC (2011) Evolutionary criteria outperform operational approaches in producing ecologically relevant fungal species inventories. Mol Ecol 20:655–666
Pringle A, Barron E, Sartor K, Wares J (2011) Fungi and the Anthropocene: biodiversity discovery in an epoch of loss. Fungal Ecol 4:121–123
Pringle A, Bever JD (2002) Divergent phenologies may facilitate the coexistence of arbuscular mycorrhizal fungi in a North Carolina grassland. Am J Bot 89:1439–1446
Robinson-Boyer L, Grzyb I, Jeffries P (2009) Shifting the balance from qualitative to quantitative analysis of arbuscular mycorrhizal communities in field soils. Fungal Ecol 2:1–9
Rosendahl S (2008) Communities, populations and individuals of arbuscular mycorrhizal fungi. New Phytol 178:253–266
Sanders IR (2004) Plant and arbuscular mycorrhizal fungal diversity — are we looking at the relevant levels of diversity and are we using the right techniques? New Phytol 164:415–418
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246
Schüßler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421
Schüßler A, Walker C (2010) The Glomeromycota: a species list with new families and new genera. URL: http://www.amf-phylogeny.com. Published in December 2010 in libraries at The Royal Botanic Garden Edinburgh, The Royal Botanic Garden Kew, Botanische Staatssammlung Munich, and Oregon State University. Electronic copy freely available online at http://www.amf-phylogeny.com. Printed copy of available under ISBN-13: 978–1466388048, ISBN-10: 1466388048. Arthur Schüßler and Christopher Walker, Gloucester
Simon L, Lalonde M, Bruns TD (1992) Specific amplification of 18S fungal ribosomal genes from VA endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58:291–295
Smith SE, Read DJ (2008) Mycorrhizal symbiosis. Academic Press, Amsterdam
Stockinger H, Krüger M, Schüßler A (2010) DNA barcoding of arbuscular mycorrhizal fungi. New Phytol 187:461–474
Stürmer SL (2012) A history of the taxonomy and systematics of arbuscular mycorrhizal fungi belonging to the phylum Glomeromycota. Mycorrhiza 22:247–258
Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Bahram M, Bechem E, Chuyong G, Kõljalg U (2010) 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188:291–301
Thiéry O, Moora M, Vasar M, Zobel M, Öpik M (2013) Inter- and intrasporal nuclear ribosomal gene sequence variation within one isolate of arbuscular mycorrhizal fungus, Diversispora sp. Symbiosis. doi:10.1007/s13199-012-0212-0
Treseder KK, Cross A (2006) Global distributions of arbuscular mycorrhizal fungi. Ecosystems 9:305–316
Turrini A, Giovannetti M (2012) Arbuscular mycorrhizal fungi in national parks, nature reserves and protected areas worldwide: a strategic perspective for their in situ conservation. Mycorrhiza 22:81–97
van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310
Walker C, Vestberg M, Demircik F, Stockinger H, Saito M, Sawaki H, Nishmura I, Schüßler A (2007) Molecular phylogeny and new taxa in the Archaeosporales (Glomeromycota): Ambispora fennica gen. sp. nov., Ambisporaceae fam. nov., and emendation of Archaeospora and Archaeosporaceae. Mycol Res 111:137–153
Wubet T, Weiß M, Kottke I, Teketay D, Oberwinkler F (2006) Phylogenetic analysis of nuclear small subunit rDNA sequences suggests that the endangered African Pencil Cedar, Juniperus procera, is associated with distinct members of Glomeraceae. Mycol Res 110:1059–1069
Yang HS, Zang YY, Yuan YG, Tang JJ, Chen X (2012) Selectivity by host plants affects the distribution of arbuscular mycorrhizal fungi: evidence from ITS rDNA sequence metadata. BMC Evol Biol 12:50
Young JPW (2012) A molecular guide to the taxonomy of arbuscular mycorrhizal fungi. New Phytol 193:823–826
Acknowledgments
Elise Vanatoa assisted with laboratory procedures. We thank the numerous colleagues and institutions who offered local support and contributed to field work: Christina Birnbaum, Helge Bruelheide, Jeff Cole, Karen J. Esler, Evelina Facelli, Lars Götzenberger, Andres Koppel, Eliane Louisanna, Eric Marcon, John Morgan, Jodi N. Price, Marina Semchenko, Sawarng Sitawan, Elise Vanatoa, Wang Wei, CENAREST (Gabon) for granting us permission to carry out research in Gabon and the ANPN for allowing us to carry out fieldwork in the national park of Monts de Cristal, Gutianshan Natural Reserve (China), Western Cape Nature Conservation Board (South Africa), and Wildlife Conservation, Development and Extension Station, Phitsanulok (Thailand). Some of the voucher specimens of AM host plants are preserved in the herbarium of Botanical and Mycological Museum (TU), Natural History Museum of University of Tartu. We are grateful to Herbier de Guyane (CAY) and in particular Dr. Jean-Jacques de Granville for kindly helping with plant identification. This research was funded by grants from the Estonian Science Foundation (9050, 9157, 7738), targeted financing (SF0180098s08), the European Regional Development Fund (Center of Excellence FIBIR), the Parrot project (14541QM) and the Hubert Curien Partnership between France and Estonia.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
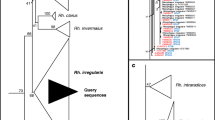
Fig. S1
Maximum clade credibility tree of Glomeromycota virtual taxa (VT), inferred using Bayesian phylogenetic analysis of small subunit rRNA gene sequences. Posterior probabilities (when >0.5) for nodes are shown. Glomeromycota nomenclature according to Schüßler et al. (2001) including modifications up to 2010, Schüßler and Walker (2010) and Oehl et al. (2011c), respectively, is shown. Family nomenclature is given in the same order, separated by slashes, unless the family names remain unchanged; in which case only one family name is given. VT detected in this study are highlighted in bold type, including novel VT which are coded as MO-xx; the sequencing approach used for each sequence is indicated (Sanger/454). Type sequences of existing VT are indicated with “Type”
Rights and permissions
About this article
Cite this article
Öpik, M., Zobel, M., Cantero, J.J. et al. Global sampling of plant roots expands the described molecular diversity of arbuscular mycorrhizal fungi. Mycorrhiza 23, 411–430 (2013). https://doi.org/10.1007/s00572-013-0482-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-013-0482-2