Abstract
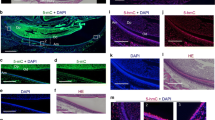
Tissue-specific gene expression is subjected to epigenetic and genetic regulation. Posttranslational modifications of histone tails alter the accessibility of nuclear proteins to DNA, thus affecting the activity of the regulatory complex of nuclear proteins. Methylation at histone 3 lysine 9 (H3K9) is a crucial modification that affects gene expression and cell differentiation. H3K9 is known to have 0–3 methylation states, and these four methylated states are determined by the expression of sets of histone methyltransferases. During development, teeth are formed through mutual interactions between the mesenchyme and epithelium via a process that is subjected to the epigenetic regulation. In this study, we examined the expression of all H3K9 methyltransferases (H3K9MTases) during mouse tooth development. We found that four H3K9MTases—G9a, Glp, Prdm2, and Suv39h1—were highly expressed in the tooth germ, with expression peaks at around embryonic days 16.5 and 17.5 in mice. Immunohistochemical and in situ hybridization analyses revealed that all four H3K9MTases were enriched in the mesenchyme more than in the epithelium. Substrates of H3K9MTases, H3K9me1, H3K9me2, and H3K9me3 were also enriched in the mesenchyme. Taken together, these data suggested that coordinated expression of four H3K9MTases in the dental mesenchyme might play important roles in tooth development.





Similar content being viewed by others
Abbreviations
- H3K9me1:
-
Monomethylated lysine 9 at histone H3
- H3K9me2:
-
Dimethylated lysine 9 at histone H3
- H3K9me3:
-
Trimethylated lysine 9 at histone H3
- H3K9MTase:
-
H3K9 methyltransferase
References
Alappat S, Zhang ZY, Chen YP (2003) Msx homeobox gene family and craniofacial development. Cell Res 13:429–442
Black JC, Van Rechem C, Whetstine JR (2012) Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48:491–507
Blackburn ML, Chansky HA et al (2003) Genomic structure and expression of the mouse ESET gene encoding an ERG-associated histone methyltransferase with a SET domain. Biochim Biophys Acta 1629:8–14
Chen ZX, Riggs AD (2011) DNA methylation and demethylation in mammals. J Biol Chem 286:18347–18353
Chen Y, Bei M et al (1996) Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development 122:3035–3044
Chen X, Skutt-Kakaria K et al (2012) G9a/GLP-dependent histone H3K9me2 patterning during human hematopoietic stem cell lineage commitment. Genes Dev 26:2499–2511
Dassule HR, McMahon AP (1998) Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol 202:215–227
Dodge E, Kang K et al (2004) Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24:2478–2486
Fritsch L, Robin P et al (2010) A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37:46–56
Ideno H, Shimada A et al (2013) Predominant expression of H3K9 methyltransferases in prehypertrophic and hypertrophic chondrocytes during mouse growth plate cartilage development. Gene Expr Patterns 13:84–90
Ideno H, Shimada A et al (2014) Search for conditions to detect epigenetic marks and nuclear proteins in immunostaining of the testis and cartilage. J Histol 2014, Art ID 658293. doi:10.1155/2014/658293
Jernvall J, Thesleff I (2012) Tooth shape formation and tooth renewal: evolving with the same signals. Development 139:3487–3497
Jia S, Zhou J et al (2013) Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development 140:423–432
Jones PA, Liang G (2009) Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10:805–811
Kratochwil K, Galceran J et al (2002) FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(-/-) mice. Genes Dev 16:3173–3185
Mozzetta C, Pontis J et al (2014) The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell 53:277–289
Nifuji A, Noda M (1999) Coordinated expression of noggin and bone morphogenetic proteins (BMPs) during early skeletogenesis and induction of noggin expression by BMP-7. J Bone Miner Res 14:2057–2066
Nifuji A, Ideno H et al (2010) Nemo-like kinase (NLK) expression in osteoblastic cells and suppression of osteoblastic differentiation. Exp Cell Res 316:1127–1136
O’Carroll D, Scherthan H et al (2000) Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol 20:9423–9433
Oh ST, Kim KB et al (2014) H3K9 histone methyltransferase G9a-mediated transcriptional activation of p21. FEBS Lett 588:685–691
Peters AH, O’Carroll D et al (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337
Purcell DJ, Khalid O et al (2012) Recruitment of coregulator G9a by Runx2 for selective enhancement or suppression of transcription. J Cell Biochem 113:2406–2414
Saadi I, Das P et al (2013) Msx1 and Tbx2 antagonistically regulate Bmp4 expression during the bud-to-cap stage transition in tooth development. Development 140:2697–2702
Shinkai Y, Tachibana M (2011) H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 25:781–788
Steele-Perkins G, Fang W et al (2001) Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev 15:2250–2262
Tachibana M, Sugimoto K, Fukushima T, Shinkai Y (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276:25309–25317
Tachibana M, Sugimoto K et al (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16:1779–1791
Tachibana M, Ueda J et al (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19:815–826
Tachibana M, Matsumura Y et al (2008) G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 27:2681–2690
Tong X, Zhang D et al (2013) Recruitment of histone methyltransferase G9a mediates transcriptional repression of Fgf21 gene by E4BP4 protein. J Biol Chem 288:5417–5425
Tucker A, Sharpe P (2004) The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet 5:499–508
Yun M, Wu J, Workman JL, Li B (2011) Readers of histone modifications. Cell Res 21:564–578
Acknowledgments
This study was supported by grants-in-aid funding for scientific research (JSPS KAKENHI Grant Numbers 22390344, 23659862, 25670784 to A.N.).
Conflict of interest
There is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kamiunten, T., Ideno, H., Shimada, A. et al. Coordinated expression of H3K9 histone methyltransferases during tooth development in mice. Histochem Cell Biol 143, 259–266 (2015). https://doi.org/10.1007/s00418-014-1284-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-014-1284-0