Abstract
Purpose
To develop a population pharmacokinetic (PK) model for HM781-36 (poziotinib) and its metabolites in cancer patients.
Methods
Blood samples were collected from three phase I studies in which fifty-two patients received oral HM781-36B tablets (0.5–32 mg) once daily for 2 weeks, and another 20 patients received oral HM781-36B tablets (12, 16, 18, 24 mg) in fasting (12 patients) or fed (eight patients) state once daily for 4 weeks. Nonlinear mixed effect modeling was employed to develop the population pharmacokinetic model.
Results
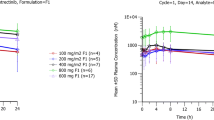
HM781-36 PK was ascribed to a two-compartment model and HM781-36-M1/-M2 PK to one-compartment model. HM781-36 oral absorption was characterized by first-order input (absorption rate constant: 1.45 ± 0.23 h−1). The central volume of distribution (185 ± 12.7 L) was influenced significantly by body weight. The absorption rate constant was influenced by food. The typical HM781-36 apparent clearance was 34.5 L/h (29.4 %CV), with an apparent peripheral volume of distribution of 164 L (53.5 %CV). Other covariates did not significantly further explain the PKs of HM781-36.
Conclusions
The proposed model suggests that HM781-36 PKs are consistent across most solid tumor types, and that the absorption process of HM781-36 is affected by the fed state before dosing. HM781-36 PKs are not complicated by patient factors, other than body weight.






Similar content being viewed by others
References
Wells A (1999) EGF receptor. Int J Biochem Cell Biol 31(6):637–643
Ritter CA, Arteaga CL (2003) The epidermal growth factor receptor-tyrosine kinase: a promising therapeutic target in solid tumors. Semin Oncol 30(1 Suppl 1):3–11
Beal SL (2001) Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn 28(5):481–504
Beal SL, Sheiner LB, Boeckmann AJ (1989) 2006 NONMEM Users Guide: Part I-VII. Icon Development Solutions, Ellicott City
Keizer RJ, Karlsson MO, Hooker A (2013) Modeling and simulation workbench for NONMEM: tutorial on Pirana, PsN, and Xpose. CPT Pharmacomet Syst Pharmacol 2:e50
Lindbom L, Pihlgren P, Jonsson EN (2005) PsN-Toolkit—a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Programs Biomed 79(3):241–257
Bergstrand M et al (2011) Prediction-corrected visual predictive checks for diagnosing nonlinear mixed-effects models. AAPS J 13(2):143–151
Jonsson EN, Karlsson MO (1999) Xpose—an S-PLUS based population pharmacokinetic/pharmacodynamic model building aid for NONMEM. Comput Methods Programs Biomed 58(1):51–64
Ette EI (1997) Stability and performance of a population pharmacokinetic model. J Clin Pharmacol 37(6):486–495
Ette EI, Onyiah LC (2002) Estimating inestimable standard errors in population pharmacokinetic studies: the bootstrap with Winsorization. Eur J Drug Metab Pharmacokinet 27(3):213–224
Parke J, Holford NH, Charles BG (1999) A procedure for generating bootstrap samples for the validation of nonlinear mixed-effects population models. Comput Methods Programs Biomed 59(1):19–29
Yano Y, Beal SL, Sheiner LB (2001) Evaluating pharmacokinetic/pharmacodynamic models using the posterior predictive check. J Pharmacokinet Pharmacodyn 28(2):171–192
Smith BP et al (2000) Confidence interval criteria for assessment of dose proportionality. Pharm Res 17(10):1278–1283
Scheffler M et al (2011) Clinical pharmacokinetics of tyrosine kinase inhibitors: focus on 4-anilinoquinazolines. Clin Pharmacokinet 50(6):371–403
Bello CL et al (2013) A phase I, open-label, mass balance study of [(14)C] dacomitinib (PF-00299804) in healthy male volunteers. Cancer Chemother Pharmacol 72(2):379–385
Karlsson MO, Sheiner LB (1993) The importance of modeling interoccasion variability in population pharmacokinetic analyses. J Pharmacokinet Biopharm 21(6):735–750
Acknowledgments
This study was sponsored by Hanmi Pharm., Co. Ltd., Seoul, Korea. The authors are grateful to the Clinical Research, Drug Metabolism and Pharmacokinetics, and Chemical Structure Analysis teams of Hanmi Pharm.
Conflict of interest
Jin-A Jung and Tae Hun Song are employees of Hanmi Pharm., Co. Ltd. The authors warrant that they have no other conflict of interest regarding the contents of this article.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1
HM781-36 Pharmacokinetic model (parent molecule only).
where all parameters are as defined in the text and individual PK parameters are denoted by the subscript i.
Appendix 2
Combined HM781-36 and HM781-36-M1/-M2 Pharmacokinetic parent-metabolite model.
where all parameters are as defined in the text, and individual PK parameters are denoted by the subscript i.
Appendix 3
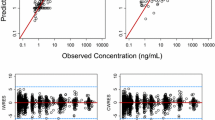
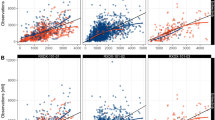
See Table 4 and Figs. 4, 5 and 6.
Rights and permissions
About this article
Cite this article
Noh, YH., Lim, HS., Jung, JA. et al. Population pharmacokinetics of HM781-36 (poziotinib), pan-human EGF receptor (HER) inhibitor, and its two metabolites in patients with advanced solid malignancies. Cancer Chemother Pharmacol 75, 97–109 (2015). https://doi.org/10.1007/s00280-014-2621-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-014-2621-7