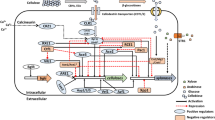
Abstract
Filamentous fungi produce cellulolytic and hemicellulolytic enzymes in response to small inducer molecules liberated from cellulosic biomass. Enzyme production is mainly regulated at the level of transcription. The first transcription factor identified as being involved in cellulosic biomass degradation was XlnR, which mediates d-xylose-triggered induction of xylanolytic and cellulolytic genes in Aspergillus. XlnR has played the leading role for over a decade in studies aimed at clarification of gene regulation related to cellulosic biomass degradation. Very recently, several new transcription factors were identified, namely Clr-1/2 in Neurospora; ManR, McmA, and ClbR in Aspergillus; and BglR in Trichoderma, all of which participate in the regulation of cellulolytic and/or hemicellulolytic enzyme production. Furthermore, as well as the carbon sources available, other factors such as light signaling and anti-sense RNA accumulation have been shown to contribute to this regulation. Here, we review the recent advancements demonstrating that multiple factors coordinately regulate the expression of cellulosic biomass degrading enzyme genes.

Similar content being viewed by others
References
Alper H, Moxley J, Nevoigt E, Fink G, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314(5805):1565–1568
Alper H, Stephanopoulos G (2007) Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng 9(3):258–267
Aro N, Pakula T, Penttila M (2005) Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev 29(4):719–739
Baker CR, Hanson-Smith V, Johnson AD (2013) Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science 342(6154):104–108
Battaglia E, Hansen SF, Leendertse A, Madrid S, Mulder H, Nikolaev I, de Vries RP (2011a) Regulation of pentose utilisation by arar, but not xlnr, differs in Aspergillus nidulans and Aspergillus niger. Appl Microbiol Biotechnol 91(2):387–397
Battaglia E, Visser L, Nijssen A, van Veluw G, Wosten H, de Vries R (2011b) Analysis of regulation of pentose utilisation in Aspergillus niger reveals evolutionary adaptations in Eurotiales. Stud Mycol(69):31-38
Berlin A, Maximenko V, Gilkes N, Saddler J (2007) Optimization of enzyme complexes for lignocellulose hydrolysis. Biotechnol Bioeng 97(2):287–296
Bernreiter A, Ramon A, Fernandez-Martinez J, Berger H, Araujo-Bazan L, Espeso EA, Pachlinger R, Gallmetzer A, Anderl I, Scazzocchio C, Strauss J (2007) Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol Cell Biol 27(3):791–802
Brunner K, Lichtenauer AM, Kratochwill K, Delic M, Mach RL (2007) Xyr1 regulates xylanase but not cellulase formation in the head blight fungus Fusarium graminearum. Curr Genet 52(5–6):213–220
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103(27):10352–10357
Coradetti S, Craig J, Xiong Y, Shock T, Tian C, Glass N (2012) Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci U S A 109(19):7397–7402
Coradetti ST, Xiong Y, Glass NL (2013) Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2(4):595–609
Coyle SM, Flores J, Lim WA (2013) Exploitation of latent allostery enables the evolution of new modes of MAP kinase regulation. Cell 154(4):875–887
Darieva Z, Clancy A, Bulmer R, Williams E, Pic-Taylor A, Morgan B, Sharrocks A (2010) A competitive transcription factor binding mechanism determines the timing of late cell cycle-dependent gene expression. Mol Cell 38(1):29–40
de Souza WR, de Gouvea PF, Savoldi M, Malavazi I, de Souza Bernardes LA, Goldman MH, de Vries RP, de Castro Oliveira JV, Goldman GH (2011) Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotech Biofuels 4:40
Delmas S, Pullan ST, Gaddipati S, Kokolski M, Malla S, Blythe MJ, Ibbett R, Campbell M, Liddell S, Aboobaker A, Tucker GA, Archer DB (2012) Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing. PLoS Genet 8(8):e1002875
Endo Y, Yokoyama M, Morimoto M, Shirai K, Chikamatsu G, Kato N, Tsukagoshi N, Kato M, Kobayashi T (2008) Novel promoter sequence required for inductive expression of the Aspergillus nidulans endoglucanase gene eglA. Biosci Biotechnol Biochem 72(2):312–320
Goosen T, Bloemheuvel G, Gysler C, Debie D, Vandenbroek H, Swart K (1987) Transformation of Aspergillus niger using the homologous orotidine-5′-phosphate-decarboxylase gene. Curr Genet 11(6–7):499–503
Hasper A, Trindade L, van der Veen D, van Ooyen A, de Graaff L (2004) Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiology 150:1367–1375
Holmberg CI, Tran SE, Eriksson JE, Sistonen L (2002) Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci 27(12):619–627
Hrmova M, Petrakova E, Biely P (1991) Induction of cellulose-degrading and xylan-degrading enzyme-systems in Aspergillus terreus by homodisaccharides and heterodisaccharides composed of glucose and xylose. J Gen Microbiol 137:541–547
Johnston M (1987) Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature 328(6128):353–355
Karaffa L, Fekete E, Gamauf C, Szentirmai A, Kubicek CP, Seiboth B (2006) D-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology 152(Pt 5):1507–1514
Kawai T, Nakazawa H, Ida N, Okada H, Tani S, Sumitani J, Kawaguchi T, Ogasawara W, Morikawa Y, Kobayashi Y (2012) Analysis of the saccharification capability of high-functional cellulase JN11 for various pretreated biomasses through a comparison with commercially available counterparts. J Ind Microbiol Biotechnol 39(12):1741–1749
Keegan L, Gill G, Ptashne M (1986) Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231(4739):699–704
Kubicek CP, Messner R, Gruber F, Mach R, Kubicek-Pranz EM (1993) The Trichoderma cellulase regulatory puzzle; from the interior life of a secretory fungus. Enzyme Microb Technol 15(2):90–99
Kubicek CP, Penttilä ME (1998) Regulation of production of plant polysaccharide degrading enzymes by Trichoderma. In: Harman GE, Kubicek CP (eds) Trichoderma and Gliocaldium. Enzymes biological control and commercial applications edn, vol 2. Taylor and Francis, London, pp 49–72
Kumar PR, Yu Y, Sternglanz R, Johnston SA, Joshua-Tor L (2008) NADP regulates the yeast GAL induction system. Science 319(5866):1090–1092
Kunitake E, Tani S, Sumitani J, Kawaguchi T (2011) Agrobacterium tumefaciens-mediated transformation of Aspergillus aculeatus for insertional mutagenesis. AMB Express 1(1):46
Kunitake E, Tani S, Sumitani J, Kawaguchi T (2013) A novel transcriptional regulator, ClbR, controls the cellobiose- and cellulose-responsive induction of cellulase and xylanase genes regulated by two distinct signaling pathways in Aspergillus aculeatus. Appl Microbiol Biotechnol 97(5):2017–2028
Kurasawa T, Yachi M, Suto M, Kamagata Y, Takao S, Tomita F (1992) Induction of cellulase by gentiobiose and its sulfur-containing analog in Penicillium purpurogenum. Appl Environ Microbiol 58(1):106–110
Lan C, Lee HC, Tang S, Zhang L (2004) A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J Biol Chem 279(26):27607–27612
Lapidot M, Pilpel Y (2006) Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep 7(12):1216–1222
Lavy T, Kumar PR, He H, Joshua-Tor L (2012) The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation. Genes Dev 26(3):294–303
Li Y, Chen G, Liu W (2010) Multiple metabolic signals influence GAL gene activation by modulating the interaction of Gal80p with the transcriptional activator Gal4p. Mol Microbiol 78(2):414–428
Liu G, Zhang L, Wei X, Zou G, Qin Y, Ma L, Li J, Zheng H, Wang S, Wang C, Xun L, Zhao G, Zhou Z, Qu Y (2013) Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS One 8(2)
Lockington R, Rodbourn L, Barnett S, Carter C, Kelly J (2002) Regulation by carbon and nitrogen sources of a family of cellulases in Aspergillus nidulans. Fungal Genet Biol 37(2):190–196
Ma J, Ptashne M (1987) A new class of yeast transcriptional activators. Cell 51(1):113–119
Makita T, Katsuyama Y, Tani S, Suzuki H, Kato N, Todd RB, Hynes MJ, Tsukagoshi N, Kato M, Kobayashi T (2009) Inducer-dependent nuclear localization of a Zn(II)2Cys6 transcriptional activator, AmyR, in Aspergillus nidulans. Biosci Biotechnol Biochem 73(2):391–399
Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM (2001) Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell 104(5):675–686
Mandels M, Parrish FW, Reese ET (1962) Sophorose as an inducer of cellulase in Trichoderma viride. J Bacteriol 83:400–408
Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EG, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, de Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barabote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26(5):553–60
Marui J, Kitamoto N, Kato M, Kobayashi T, Tsukagoshi N (2002) Transcriptional activator, AoXlnR, mediates cellulose-inductive expression of the xylanolytic and cellulolytic genes in Aspergillus oryzae. FEBS Lett 528(1–3):279–282
Messenguy F, Dubois E (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316:1–321
Morikawa Y, Ohashi T, Mantani O, Okada H (1995) Cellulase induction by lactose in Trichoderma reesei PC-3-7. Appl Microbiol Biotechnol 44(1–2):106–111
Murakoshi Y, Makita T, Kato M, Kobayashi T (2012) Comparison and characterization of alpha-amylase inducers in Aspergillus nidulans based on nuclear localization of AmyR. Appl Microbiol Biotechnol 94(6):1629–1635
Nakazawa H, Kawai T, Ida N, Shida Y, Kobayashi Y, Okada H, Tani S, Sumitani J, Kawaguchi T, Morikawa Y, Ogasawara W (2012) Construction of a recombinant Trichoderma reesei strain expressing Aspergillus aculeatus beta-glucosidase 1 for efficient biomass conversion. Biotechnol Bioeng 109(1):92–99
Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci U S A 101(33):12248–12253
Nitta M, Furukawa T, Shida Y, Mori K, Kuhara S, Morikawa Y, Ogasawara W (2012) A new Zn(II)2Cys6-type transcription factor BglR regulates beta-glucosidase expression in Trichoderma reesei. Fungal Genet Biol 49(5):388–397
Noguchi Y, Sano M, Kanamaru K, Ko T, Takeuchi M, Kato M, Kobayashi T (2009) Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl Microbiol Biotechnol 85(1):141–154
Noguchi Y, Tanaka H, Kanamaru K, Kato M, Kobayashi T (2011) Xylose triggers reversible phosphorylation of XlnR, the fungal transcriptional activator of xylanolytic and cellulolytic genes in Aspergillus oryzae. Biosci Biotechnol Biochem 75(5):953–959
Ogawa M, Kobayashi T, Koyama Y (2012) ManR, a novel Zn(II)2Cys6 transcriptional activator, controls the beta-mannan utilization system in Aspergillus oryzae. Fungal Genet Biol 49(12):987–995
Ogawa M, Kobayashi T, Koyama Y (2013) ManR, a transcriptional regulator of the beta-mannan utilization system, controls the cellulose utilization system in Aspergillus oryzae. Biosci Biotechnol Biochem 77(2):426–429
Poulou M, Bell D, Bozonelos K, Alexiou M, Gavalas A, Lovell-Badge R, Remboutsika E (2010) Development of a chromosomally integrated metabolite-inducible Leu3p-alpha-IPM “off-on” gene switch. PLoS One 5(8):e12488
Schmoll M, Tian C, Sun J, Tisch D, Glass N (2012) Unravelling the molecular basis for light modulated cellulase gene expression—the role of photoreceptors in Neurospora crassa. BMC Genomics 13:127
Schuster A, Tisch D, Seidl-Seiboth V, Kubicek CP, Schmoll M (2012) Roles of protein kinase a and adenylate cyclase in light-modulated cellulase regulation in Trichoderma reesei. Appl Environ Microbiol 78(7):2168–2178
Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, Sauer DG, Smith KM, Baker SE, Freitag M, Kubicek CP (2012) The putative protein methyltransferase Lae1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol 84(6):1150–1164
Shore P, Sharrocks A (1995) The MADS-box family of transcription factors. Eur J Biochem 229(1):1–13
Sil AK, Alam S, Xin P, Ma L, Morgan M, Lebo CM, Woods MP, Hopper JE (1999) The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol Cell Biol 19(11):7828–7840
Somerville C (2006) The billion-ton biofuels vision. Science 312(5778):1277–1277
Stricker AR, Grosstessner-Hain K, Wurleitner E, Mach RL (2006) Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot Cell 5(12):2128–2137
Stricker AR, Mach RL, de Graaff LH (2008) Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl Microbiol Biotechnol 78(2):211–220
Stricker AR, Steiger MG, Mach RL (2007) Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett 581(21):3915–3920
Sun J, Glass NL (2011) Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One 6(9):e25654
Sze JY, Woontner M, Jaehning JA, Kohlhaw GB (1992) In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science 258(5085):1143–1145
Tani S, Kanamasa S, Sumitani J, Arai M, Kawaguchi T (2012) XlnR-independent signaling pathway regulates both cellulase and xylanase genes in response to cellobiose in Aspergillus aculeatus. Curr Genet 58(2):93–104
Tilburn J, Sarkar S, Widdick D, Espeso E, Orejas M, Mungroo J, Penalva M, Arst H (1995) The Aspergillus PacC zinc-finger transcription factor mediates regulation of both acid-expressed and alkaline-expressed genes by ambient pH. EMBO J 14(4):779–790
Todd R, Andrianopoulos A (1997) Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol 21(3):388–405
van Peij N, Gielkens M, de Vries R, Visser J, de Graaff L (1998) The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol 64(10):3615–3619
Wang D, Zheng F, Holmberg S, Kohlhaw GB (1999) Yeast transcriptional regulator Leu3p. Self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. J Biol Chem 274(27):19017–19024
Yamakawa Y, Endo Y, Li N, Yoshizawa M, Aoyama M, Watanabe A, Kanamaru K, Kato M, Kobayashi T (2013) Regulation of cellulolytic genes by McmA, the SRF-MADS box protein in Aspergillus nidulans. Biochem Biophys Res Commun 431(4):777–782
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tani, S., Kawaguchi, T. & Kobayashi, T. Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Appl Microbiol Biotechnol 98, 4829–4837 (2014). https://doi.org/10.1007/s00253-014-5707-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5707-6