Abstract
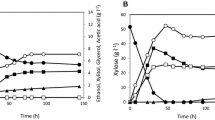
Recombinant yeast strains highly tolerant to formic acid during xylose fermentation were constructed. Microarray analysis of xylose-fermenting Saccharomyces cerevisiae strain overexpressing endogenous xylulokinase in addition to xylose reductase and xylitol dehydrogenase from Pichia stipitis revealed that upregulation of formate dehydrogenase genes (FDH1 and FDH2) was one of the most prominent transcriptional events against excess formic acid. The quantification of formic acid in medium indicated that the innate activity of FDH was too weak to detoxify formic acid. To reinforce the capability for formic acid breakdown, the FDH1 gene was additionally overexpressed in the xylose-metabolizing recombinant yeast. This modification allowed the yeast to rapidly decompose excess formic acid. The yield and final ethanol concentration in the presence of 20 mM formic acid is as essentially same as that of control. The fermentation profile also indicated that the production of xylitol and glycerol, major by-products in xylose fermentation, was not affected by the upregulation of FDH activity.




Similar content being viewed by others
References
Almeida JRM, Modig T, Petersson A, Hahn-Hägerdal B, Lidén G, Gorwa-Grauslund MF (2007) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349
Amore R, Kötter P, Küster C, Ciriacy M, Hollenberg CP (1991) Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene 109(1):89–97
Chu BC, Lee H (2007) Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv 25(5):425–441
Fu TF, Rife JP, Schirch V (2001) The role of serine hydroxymethyltransferase isozymes in one-carbon metabolism in MCF-7 cells as determined by 13C NMR. Arch Biochem Biophys 393(1):42–50
Geertman JM, van Dijken JP, Pronk JT (2006a) Engineering NADH metabolism in Saccharomyces cerevisiae: formate as an electron donor for glycerol production by anaerobic, glucose-limited chemostat cultures. FEMS Yeast Res 6(8):1193–1203
Geertman JM, van Maris AJ, van Dijken JP, Pronk JT (2006b) Physiological and genetic engineering of cytosolic redox metabolism in Saccharomyces cerevisiae for improved glycerol production. Metab Eng 8(6):532–542
Hirasawa T, Yoshikawa K, Nakakura Y, Nagahisa K, Furusawa C, Katakura Y, Shimizu H, Shioya S (2007) Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J Biotechnol 131(1):34–44
Ishii J, Izawa K, Matsumura S, Wakamura K, Tanino T, Tanaka T, Ogino C, Fukuda H, Kondo A (2009) A simple and immediate method for simultaneously evaluating expression level and plasmid maintenance in yeast. J Biochem 145(6):701–708
Katahira S, Mizuike A, Fukuda H, Kondo A (2006) Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol 72(6):1136–1143
Katahira S, Ito M, Takema H, Fujita Y, Tanino T, Tanaka T, Fukuda H, Kondo A (2008) Improvement of ethanol productivity during xylose and glucose co-fermentation by xylose-assimilating S. cerevisiae via expression of glucose transporter Sut1. Enzyme Microb Technol 43:115–119
Keating JD, Panganiban C, Mansfield SD (2006) Tolerance and adaptation of ethanologenic yeasts to lignocellulosic inhibitory compounds. Biotechnol Bioeng 93(6):1196–1206
Kennedy CJ, Boyle PM, Waks Z, Silver PA (2009) Systems-level engineering of nonfermentative metabolism in yeast. Genetics 183(1):385–397
Kötter P, Ciriacy M (1993) Xylose fermentation by Saccharomyces cerevisiae. Appl Microb Biotechnol 38(6):776–783
Kötter P, Amore R, Hollenberg CP, Ciriacy M (1990) Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet 18(6):493–500
Li BZ, Yuan YJ (2010) Transcriptome shifts in response to furfural and acetic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 86(6):1915–1924
Lin Y, Tanaka S (2006) Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol 69(6):627–642
Matsushika A, Watanabe S, Kodaki T, Makino K, Sawayama S (2008) Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP+-dependent xylitol dehydrogenase, and xylulokinase. J Biosci Bioeng 105(3):296–299
Matsushika A, Inoue H, Kodaki T, Sawayama S (2009) Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol 84(1):37–53
Milliken CE, May HD (2007) Sustained generation of electricity by the spore-forming, Gram-positive, Desulfitobacterium hafniense strain DCB2. Appl Microbiol Biotechnol 73(5):1180–1189
Negro MJ, Manzanares P, Ballesteros I, Oliva JM, Cabañas A, Ballesteros M (2003) Hydrothermal pretreatment conditions to enhance ethanol production from poplar biomass. Appl Biochem Biotechnol 105(1–3):87–100
Overkamp KM, Kötter P, van der Hoek R, Schoondermark-Stolk S, Luttik MA, van Dijken JP, Pronk JT (2002) Functional analysis of structural genes for NAD+-dependent formate dehydrogenase in Saccharomyces cerevisiae. Yeast 19(6):509–520
Palmqvist E, Hahn-Hägerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 74(1):25–33
Petersson A, Almeida JR, Modig T, Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF, Lidén G (2006) A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23(6):455–464
Schüller C, Mamnun YM, Mollapour M, Krapf G, Schuster M, Bauer BE, Piper PW, Kuchler K (2004) Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell 15(2):706–720
Sonderegger M, Jeppsson M, Hahn-Hägerdal B, Sauer U (2004) Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Appl Environ Microbiol 70(4):2307–2317
Tajima M, Nogi Y, Fukusawa T (1985) Primary structure of the Saccharomyces cerevisiae GAL7 gene. Yeast 1:67–77
Tomás-Pejó E, Ballesteros M, Oliva JM, Olsson L (2010) Adaptation of the xylose fermenting yeast Saccharomyces cerevisiae F12 for improving ethanol production in different fed-batch SSF processes. J Ind Microbiol Biotechnol 37(11):1211–1220
van Maris AJA, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MAH, Wisselink HW, Scheffers WA, van Dijken JP, Pronk JT (2006) Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Leeuwenhoek 90:391–418
van Vleet JH, Jeffries TW (2009) Yeast metabolic engineering for hemicellulosic ethanol production. Curr Opin Microbiol 20:300–306
Verduyn C, Postma E, Scheffers WA, Van Dijken JP (1992) Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8(7):501–517
Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H (2009) Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 9(1):32–44
Acknowledgments
This work was financed by the New Energy and Industrial Technology Development Organization (NEDO), Japan, and Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hasunuma, T., Sung, Km., Sanda, T. et al. Efficient fermentation of xylose to ethanol at high formic acid concentrations by metabolically engineered Saccharomyces cerevisiae . Appl Microbiol Biotechnol 90, 997–1004 (2011). https://doi.org/10.1007/s00253-011-3085-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3085-x