Abstract
Actinobacteria are major producers of secondary metabolites; however, it is unclear how they are distributed in the environment. DNA was extracted from forest, pasture and cultivated soils, street sediments (dust and material in place), and sediments affected by animal activity (e.g. guano, vermicompost) and characterised with two actinobacterial and a bacterial-specific 16S rDNA primer set. Amplicons (140/156) generated with the two actinobacterial-specific and amplicons (471) generated with bacterial-specific primers were analysed. Amplicons from actinobacterial-specific primer were disproportionately actinomycetal from animal-affected (soil) samples and street sediments and either verrucomicrobial (i.e. non-actinobacterial) and from a novel non-actinomycetal actinobacterial group for soils. Actinobacterial amplified ribosomal DNA restriction analysis and terminal restriction fragment length polymorphism fingerprints clustered by land use, with cultivated soils clustering apart from uncultivated soils. Actinobacterial amplicons generated with eubacterial primers were overwhelmingly from (116/126) street sediments; acidobacterial amplicons from soils (74/75). In two street samples, >90% of clones were actinomycetal. Actinomycetes are selected in terrestrial soils and sediments by cultivation, urbanisation and animal activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most known antibiotics from bacteria are produced by Actinobacteria. Within this phylum, members of the order Actinomycetes are the major producers of antibiotics and in some cases anticancer agents and immunosuppressants. The identification of new Actinomycetes thus can play an important role in drug discovery. Whilst Actinobacteria are generally considered as soil-dwelling bacteria, it remains unclear what determines their community structure. Understanding actinobacterial distribution in the environment is important in understanding their ecological role and for pharmaceutical bioprospecting.
The distribution of soil bacterial communities does not correspond to eukaryotic biomes such as rainforest, prairie or tundra [32]. There is no consensus on how terrestrial microbial populations are distributed. Several studies have found that soil bacterial community structure is more affected by the soil properties than the vegetation that the soil supports [3, 11]. Soil texture [10], pH [8] or parent material [43] have all been suggested as the controlling factors for soil bacterial community structure.
Within the bacterial community, bacterial subgroups can be controlled by factors other than those that influence the entire community structure. For example, the proteobacterial Psychrobacter and firmicute Exiguobacterium genera are more common in soil samples from cold climates [35]. Additionally, the community structure of soil ammonium-oxidising bacteria is also controlled by climate [9]. Identifying factors which control actinobacterial community structure will thus play an important role in identifying sampling sites for bioprospecting.
Two culture-independent studies have found contrasting factors influencing actinobacterial community structure in soil. Wawrik et al. [42] compared actinobacterial-specific terminal restriction fragment length polymorphism (T-RFLP) patterns from Uzbekistan and New Jersey, USA, and found a strong biogeographic effect. Based on these results, he recommended that bioprospectors sample geographically separated sites. Lauber et al. [26], using pyrosequencing, determined that pH, rather than biogeography, controlled both actinobacterial and total bacterial community structures. These results suggest that sampling should be directed by soil pH.
Molecular methods give a culture-independent view of the Actinobacteria and have shown that non-actinomycetal actinobacterial groups such as the Rubrobacteria and Acidimicrobia are as common as the Actinomycetes in soil [17, 20]. However, the results of molecular methods are highly dependent on primer choice and extraction method. A recent study suggests that there is little overlap in the 16S sequences generated using different primers and extraction methods and that these limitations cannot be overcome by more extensive sequencing [18]. Therefore, the use of several primer systems for both fingerprinting and sequencing may give a more reliable view of community composition. In this context, sequencing is used to interpret the results of community fingerprinting rather than to characterise community structure alone.
To investigate the controlling factor for actinobacterial community structure, we applied two actinobacterial primer systems to a broad range of environmental DNA samples and applied a eubacterial primer system to a subset of soil and street sediment DNA samples. The primer sets used were the system of Heuer et al. [15], which uses a forward actinobacterial-specific 243-F primer and a eubacterial-specific reverse primer (F-Act/R-Bact); that of Monciardini et al. [30], which replaces the reverse primer with the actinobacterial-specific A3 primer (F-Act/R-Act); and the eubacterial primer system of Marchesi et al. [28], which uses 63-F and 1387-R primers. Environmental DNA was obtained from sites selected to cover a broad range of sample diversity. Samples from pasture, forest and cultivated soils on contrasting parent material (Table 1) were included, as were samples of non-soil sediments associated with humans and animals: street sediments, earthworm casts, vermicompost, beehive waste, isopoda dung and guano (Table 2). Sampling sites were located in Colombia, Canada, the Czech Republic and Siberia. Together, these samples cover a wide range of parent materials, climates and land use in diverse geographical locations. The results from our community fingerprint analyses coupled with sequencing show that overall actinobacterial community structure appears to be controlled by land use and that street sediments are enriched with Actinomycetes.
Methods
Sampling for Characterisation with Actinobacterial-Specific Primers
Soils from contrasting parent materials were sampled from areas within 20–50 km of Cali, (Colombia), Ottawa, (Canada) and České Budějovice (Czech Republic; Table 1). A single soil was sampled from the Yenisei Valley, Siberia, Russia. A medieval church garden soil from the centre of České Budějovice (garden of paradise) was also sampled. Samples were taken by driving a 5-cm-deep ring into the surface of the mineral layer of soil. Most samples were composites of five cores taken randomly from a 1-m2 area. Exceptions were the Yenisei site (Rusforest-Yenisei) and a site near České Budějovice (Czcultivated-ČeskeBudéjovice) where samples were composites of 20 cores taken from a 20 × 20-m area. As the object of sampling was to sample as many contrasting soils as possible, most sites were only sampled once. At four sites, two samples were taken about 50 m from each other (Colcultivated-Rozo 1 and 2, Colforest-Rozo 1 and 2, Colforest-Hatico 1 and 2, Czcultivated-Netolice 1 and 2).
Samples were also taken from a range of non-soil animal-associated sediments. These sediments were: earthworm casts taken from a microcosm experiment [16], bat cave guano that was heavily colonised by isopoda (Slovakia-Guano) and isopoda dung (Slovakia-Isopodadung) from the Domica cave system in Slovakia [33], manure (Czech-Manure) and vermicompost (Czech-Vermicompost) from a composting system, and soil that had received dead bees (i.e. chitin-enriched anthropogenic environment) from a hive that had been left at least 15 years on a site in the Czech Republic (Netolice-Hivewaste).
Street sediment samples were either dust from street surfaces where there might be continual transport of both sediment and bacteria or sediment taken from spaces between cobblestones or an abandoned tramline where the presence of debris (heterogeneous pieces of broken glass, screws, marbles and pottery fragments) suggested that the material had been in place for years. Street sediments were sampled in five urban areas: the suburb Goring of Worthing (UK) and the centres of České Budějovice, the Czech Republic, Groningen, the Netherlands, Paris and Brussels (Table 2).
Samples for Characterisation with Eubacterial Primers
To confirm the results obtained with actinobacterial-specific primers, two environments that gave contrasting results with actinobacterial-specific primers, soil and street sediments, were characterised with eubacterial primers. Soils were collected in either open cultivated fields (three) or under tree canopy (three). Street sediments were either street dust (three) or sediments that had collected between cobblestones or a pavement crack (three). Four soil and four street sediment DNA samples were used from the actinobacterial study. Four additional DNA extractions were performed: (1) on an acid forest soil from near České Budějovice (Czforest-Kolny) and (2) three samples from two new sampling areas: soil under woodland (Hunforest-Citadel) and sediment from a railway station pavement crack (Budapest-trainstation-fissure) were sampled in Budapest, Hungary, and street dust was sampled in the centre of Faisalabad, Pakistan (Faisalabad-streetdust-clock) by a collaborator. This additional sampling was to ensure that earlier results were not an artefact of extraction or handling.
All samples with the exception of Rusforest-Yenisei were stored at 4°C for no more than 3 days before a 1.5- 2.5-g subsample was mixed with 30 mL of 50 mM EDTA/50 mM Tris/HCl (pH 8.3) buffer, centrifuged at 6,000×g at 4°C for 30 min and the supernatant discarded before storage at −20°C. This washing step was carried out to homogenise extraction conditions and remove contaminants that might interfere with PCR. The Yenisei sample was stored for several months at −80°C before washing and DNA extraction.
Texture, pH and organic carbon content of samples were measured either in the soil analysis laboratories of the International Centre for Tropical Agriculture (CIAT) near Palmira, Colombia, the BC ASCR, v. v. i. Institute of Soil Biology, in České Budějovice, the Czech Republic, or Wageningen Agricultural University, the Netherlands (Tables 1 and 2).
DNA Extraction from Soils and Sediments
DNA was extracted from soil using a modified version of Zhou and Bruns’ [45] direct extraction method. Briefly, 2.5 mL of a 500 mM NaCl/50 mM Tris/50 mM EDTA buffer (pH 8.0) was added to soil pellets for digestion with lysozyme and proteinase K at 37°C. Lysozyme was first added to a final concentration of 5 mg/mL and samples were incubated for 1 h. Of 20% SDS, 140 μL was then added along with 1 mg of proteinase K for a further hour of digestion. Five millilitres of a 500 mM NaCl/300 mM succinic acid/10 mM ETDA (pH 5.7) buffer was then added, followed by 700 μL of 20% SDS before a 30-min incubation at 65°C. After centrifugation, the extract was CTAB cleaned before precipitation with PEG 8000. Pellets were resuspended in 240 μL TE in Eppendorf tubes before being cleaned with a 25:24:1 phenol/chloroform/isoamyl alcohol mixture and subsequently with a 24:1 chloroform/isoamyl alcohol mixture. Remaining humic acids were precipitated by adding CaCl22H2O to a final concentration of 0.4% (w/v) and incubation at 65°C for 1 h before centrifugation. Samples were then cleaned and precipitated with 3 M potassium acetate, washed with 70% ethanol and dissolved in 20 μL TE. DNA concentration was measured with a Hoefer DyNA Quant 200 Fluorometer. Final DNA yields were between 2.3 and 30.0 μg/g soil or sediment (air dry weight).
16S rDNA Primers
In the F-Act/R-Bact amplification system of Heuer et al. [15], a single forward actinobacterial-specific primer 226-243-F (5-GGATGAGCCCGCGGCCTA-3) is used in combination with the reverse bacterial primer R-1378 CGGTCTCTACAAGGCCCGGGAACG). Monciardini et al. [30] combined the 226-243-F primer used by Heuer et al. [15] with an actinobacterial-specific reverse primer, i.e. (A3R) 1414-1430-R (5-CCAGCCCCACCTTCGAC-3). The bacterial primers of Marchesi et al. [28] consist of the forward 63-F CAGGCCTAACAC ATGCAAGTC and the reverse 1387-R GGGCGGWGTGTACAAGGC primers.
All three primer systems were used for cloning and sequencing. Products from the primers of Heuer et al. were used for amplified ribosomal DNA restriction analysis (ARDRA); products from the system of Monciardini et al. were used for T-RFLP.
PCR Amplification-F-Act/R-Bact
Cycling conditions for the primers of Heuer et al. [15] were: 35 cycles of 94°C (1 min)/64°C (1 min)/72°C (2 min), followed by a final extension of 72°C for 10 min. Amplification was carried out on 10–20 ng of environmental DNA in a total volume of 50 μL. Two units of in-house Taq polymerase produced at the CIAT biotechnology laboratories was used with Sigma PCR buffer P-2192 with a final concentration of 10 mM Tris–HCl (pH 8.3), 1.5 mM MgCl2 and 0.001% gelatin. PCR took place with 0.2 mM of each dNTP and 100 nM >of each primer. PCR products were run on a 0.8% agarose gel to check for successful amplification. Three samples did not give amplicons with the F-Act/R-Bact primers: Colforest-Rozo1, Rozo-wormcastMartio1, Rozo-wormcastMartio2.
PCR Amplification—F-Act/R-Act
Cycling conditions for the primers of Monciardini et al. [30] were: 30 cycles of 94°C (30 s)/68°C (2 min)/72°C (1 min), followed by a final extension of 72°C for 10 min. Amplification was carried out on a template of 10–20 ng of environmental DNA in a total volume of 50 μL. In addition to 2 U of Roche Taq and 10× buffer (1 × concentration 10 mM Tris–HCl/50 mM KCl/1.5 mM MgCl2, pH 8.3), the PCR cocktail had the following concentrations: 2% DMSO, 0.4 mM each dNTP,0.2 mg/mL bovine serum albumin and 500 nM of each primer. The 10× Roche buffer gives a final MgCl2 concentration of 1.5 mM; this was raised to 1.825 mM. PCR products were run on a 1% agarose gel to check for successful amplification.
PCR Amplification—Bacterial Primers
Cycling conditions for the bacterial primers of Marchesi et al. [28] were: 30 cycles of 95°C (1 min)/55°C (1 min)/72°C (1.5 min), followed by a final extension of 72°C for 5 min. Amplification was carried out on 10–20 ng of environmental DNA in a total volume of 50 μL. In addition to 2 U of Roche Taq and 10× buffer (1 × concentration 10 mM Tris–HCl/50 mM KCl/1.5 mM MgCl2, pH 8.3), the PCR cocktail had the following concentrations: 2% DMSO, 0.2 mM each dNTP,0.2 mg/mL bovine serum albumin and 200 nM of each primer.
Cloning and Sequencing from F-Act/R-Bact Primer Amplicons
PCR products from 20 environmental DNA samples (11 soils and 9 animal-affected and urban sediments; Tables 1 and 2) were used for cloning with the Invitrogen TOPO TA cloning system. White colonies were picked and used for colony PCR using the T7 and SP-6 vector primers. PCR products were sent to the University of South Bohemia, the Czech Republic, for simultaneous bidirectional sequencing using a Li-Cor 4200 L automated DNA sequencer. All of the 1,100-bp amplicons of the 155 clones were sequenced.
Cloning and Sequencing from F-Act/R-Act and Bacterial Primers Amplicons
F-Act/R-Act PCR products from ten environmental DNA samples (six soils and four other sediments) and bacterial PCR products from six soils and six street sediments (Tables 1 and 2) were picked for cloning and sequencing with 16S rDNA gene-specific primers. PCR products were cleaned with QIAquick PCR purification columns before cloning using the Promega pGEM-T easy cloning system directly. Colonies were plated on X-gal and white colonies picked and used for colony PCR using the T7 and SP-6 vector primers. Sequencing reactions were carried out on the PCR products using either the universal bacterial 1406 R primer (F-Act/R-Act PCR products) or 765-F primer (bacterial PCR products) and the ABI prism Big Dye terminator sequence reaction. Sequence reaction products were precipitated and cleaned with 75% isopropanol, dried at 50°C and dissolved in 1.0 μL of a mixture of 0.72 deionised formamide/0.28 μL loading dye. The products were denatured at 95°C for 3 min, placed on ice and loaded on an ABI377 sequencer. Sequences were extracted and analysed with GeneScanTM analysis package of programmes. Five hundred to 700 bp were sequenced from each clone. Base calling was carried out in Chromas, version 2.1. 155 (F-Act/R-Act) and 520 (Bacterial) clones were sequenced at the Centre for Evolutionary and Ecological studies sequencing facility in Haren, the Netherlands. All F-Act/R-Act sequences were used. After the exclusion of chimerical and sequences that were too short to align, 471 bacterial sequences were used for further analysis.
Sequence Data analysis and Accession Numbers
All 16S rRNA gene sequences were screened for chimaeras using the CHECKCHIMERA programme of the Ribosomal Database programme, version 8.0 [6]. Fifteen sequences generated with the F-Act/R-Bact primer system and 49 sequences generated with eubacterial primers were rejected for further analysis as being either chimerical or too short for analysis. Five sequences generated with the F-Act/R-Bact primer system were either non-chimerical or chimaeras from closely related Verrucomicrobia; these sequences were used for analysis but not submitted to Genebank (CdnforestKemptH1, ColCIATH1, CdnforestKemptH2, ColforestPanceH10, NetoliceHiveH5). Sequences from both 16S rDNA actinobacterial and primer systems were aligned and phylogenetic trees were constructed using the MEGA, version 3.0 (Molecular Evolutionary Genetics Analysis) programme [23]. Clustering was carried out using the neighbour joining algorithm bootstrapping 1,000 times. ARDRA and T-RFLP bands for BLAST matches and clone sequences were simulated either using the TAP-TRFLP programme of the ribosomal database project (Release 8.0) or Bioedit.
Three Newick files using sequences from the F-Act/R-Bact, F-Act/R-Act and bacterial primers were submitted to the Unfrac site (http://bmf2.colorado.edu/unifrac/index.psp) [27] for lineage-specific analysis.
For nucleotide sequence accession numbers, sequences were deposited in Genbank, accession numbers: F-Act/R-Bact HM444620-HM444755, F-Act/R-Act GQ494151-GQ494304 and Bacterial HM444148-HM444619.
ARDRAF-Act/R-Bact
PCR products from soil/sediment DNA using the F-Act/R-Bact primers were precipitated with ethanol and a final concentration of 0.3 M potassium acetate. Pellets were washed with 70% ethanol and resuspended in 20 μL of restriction buffer E before digestion with 1 μL of TaqI (Promega). Of the restriction digest, 3–5 μL was then run at 0.5 V/cm for 2 h on a 0.7% agarose/1.5% Synergel (Diversified Biotech) gel. Gels were stained with ethidium bromide and images captured on a GenoSmart UV transilluminator. Band sizes were estimated using the Genosoft software package (VWR International, West Chester, PA).
T-RFLP F-Act/R-Act
PCR was carried out in 50 μL reaction volumes as described above using hexachloro-6-carboxyfluorescein and carboxyfluorescin-labelled forward and reverse primers, respectively. Actinobacterial PCR products were cleaned and concentrated in 20 μL of restriction buffer using Qiagen QIAquick PCR purification columns before digestion with 2 μL of HhaI (Promega). Digested PCR product (1.1–4 μL) was dried at 50°C for 5–10 min, redissolved in 1.1 μL of 55 deionised formamide/11 ROX 1000 marker (Applied Biosystems)/11 loading dye (Applied Biosystems), denatured at 95°C for 3 min, immediately placed on ice and then run on an Applied Biosystems ABI377 sequencer to generate T-RFLP patterns. T-RFLP patterns were analysed using peaks in the 81- 677-bp range. All T-RFLP patterns used had cumulative peak heights >10,000 units [2]; for eight samples, reverse patterns and for one sample the forward pattern did not meet this criterion and were not used for analysis.
Analysis of ARDRA and T-RFLP Patterns
Patterns were imported into GELCOMPAR (Applied Maths, http://www.applied-maths.com) and compared using unweighted pair group method with arithmetic mean (UPGMA) cluster analysis.
Redundancy analysis (RDA) of patterns from samples for which pH, organic matter and clay content were measured (i.e. excluding most animal-associated sediments where high organic matter contents made it impractical to determine texture) was carried out using canonical community ordination (CANOCO) as provided by Plant Research International BV, Wageningen, the Netherlands. Community similarities were shown on ordination tri-plots, with scaling focussed on inter-sample differences and species (i.e. bands) data removed for ease of presentation. A Monte Carlo permutation test based on 499 random permutations was carried out, with the null hypothesis that patterns were unrelated to land use. Land use was treated as a nominal variable with declining values for increasing intensity of use (i.e. uncultivated soils, 3; cultivated soils, 2; street sediments, 1). Organic matter, pH and clay content were treated as supplementary variables. Organic matter and clay content were log-transformed before analysis as they were expressed as percentages; species data (i.e. band information), was square root-transformed.
Results
Sequences Generated Using the Single 16S rDNA Actinobacterial-Specific Primer System (F-Act/R-Bact)
Sequencing results show that the actinobacterial-specific primers F-Act/R-Bact amplify actinobacterial and verrucomicrobial 16S rDNA. One hundred and forty 16S rDNA amplicons from the F-Act/R-Bact primer system were sequenced and more than half of these sequences (75/140) were not actinobacterial. The majority of the non-actinobacterial clones sequenced were Verrucomicrobial (71/75). These clones are from two Verrucomicrobial subdivisions: subdivision 2, renamed Spartobacteria [39], and subdivision 3. These subdivisions are the dominant Verrucomicrobia in soil [40]. Of the 71 Verrucomicrobia sequences identified, 53 were from soil samples (Electronic Supplementary Material (ESM) Supplemental data 1). Lineage-specific analysis with UniFrac confirmed that there was selection for the Verrucomicrobia in soils (P = 0.0001, Fig. 1). The remaining four non-actinobacterial sequences grouped with the AD-3 candidate division previously found in a sandy acid soil from eastern USA [44]. All four clones were from soils with pH ≤ 5.0, and all of the closest BLAST matches were from soils and sediments with pH ≤ 5.5 (ESM Supplemental data 2).
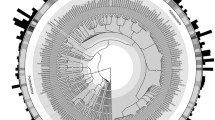
Neighbour-joining tree of 16S sequences generated with the F-Act/R-Bact primers of Heuer et al. [15]. Bootstrap values below 50 are not shown. Samples are colour coded as: forest/pasture soils (blue), insect- and earthworm-associated sediments (orange), street sediments (red). Unifrac lineage analysis was carried out on nodes A and B. For node A the P value was <0.0002 and observed/expected occurrences were for soils 53/37.7, street sediments 2/6.6 and animal affected sediments 16/26.7. For node B the P value was <0.0000 and observed/expected occurrences were for soils 17/35.1, street sediments 11/6.1 and animal affected sediments 37/28.8
The remaining 65 clones generated with the F-Act/R-Bact primers were actinobacterial. Actinobacterial clones were from Streptomycineae, Micrococcineae, Frankineae, Propionibacterineae, Pseudonocardineae and Corynebacterineae. Six clones from soil and worm gut were from a novel, non-actinomycetal. Actinobacterial group (the soil and sediment group, SOSED) is further discussed below. Actinobacterial clones were predominantly identified (48 of 65) in samples from animal-affected and urban sediments (Fig. 1), with a smaller fraction of clones (17 of 65) identified in soils. Lineage-specific analysis with UniFrac confirmed that there was selection for Actinobacteria in non-soil sediments (P = 0.0000, Fig. 1 and ESM Supplemental data 3).
Sequences Generated Using the 16S rDNA Double Actinobacterial-Specific Primers (F-Act/R-Act)
Given the poor specificity of the F-Act/R-Bact primers, samples were characterised with the more specific F-Act/R-Act primers. All clones generated with the F-Act/R-Act primers were actinobacterial. Two phylogenetic groups, a novel group (the SOSED group) and the Nocardioidaceae family constituted 79% of the clones of the library generated with the F-Act/R-Act primers (61/156 and 62/156, respectively). The SOSED group made up the majority of soil clones and the Nocardioidaceae the majority of street and animal sediment clones.
There was considerable difference in the Actinobacteria identified using the F-Act/R-Bact and F-Act/R-Act primer sets. The SOSED group and Nocardioidaceae were substantially less represented in the actinobacterial clones generated using the F-Act/R-Bact primers (6/65 and 12/65, respectively.). In addition, the Frankineae and non-mycobacterial Corynebacterineae found with the F-Act/R-Bact primer set were not detected with the F-Act/R-Act primers.
A large number of clones from the new SOSED group were identified. The closest BLAST matches to the SOSED group are from environmental clones from soils in Australia [17], Hawaii [12], Germany [13], England [7], California [24] and the Netherlands [19], as well as deep sea sediments [34], freshwater lake sediments [31], uranium mining waste [38] and a hot spring [22] (ESM Supplemental data 4). A preliminary designation of this group by İnceoğlu et al. [19] was environmental group 2; we refer to it as the SOSED (soil/sediment) group as the BLAST matches and our sequences were from marine and terrestrial soils and sediments. The vast majority of SOSED amplicons were from soil samples (58/61), and of the 79 soil amplicons, 58 were from the SOSED group. Lineage-specific analysis with UniFrac confirmed that there was selection for the SOSED group in soils (P = 0.0000, Fig. 2).
Neighbour-joining tree of 16S clones generated with the F-Act/R-Act primers of Monciardini et al. (2003). Bootstrap valules below 50 are not shown. Simulated T-RFLP values are shown for all matches were sequence length allows. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). Unifrac lineage analysis was carried out on nodes A and B. For node A, the P value was <0.0000 and observed/expected occurrences were for soils 19/43, street sediments 25/14.5 and animal-affected sediments 42/28.5. For node B, the P value was <0.0000 and observed/expected occurrences were for soils 58/29, street sediments 0/9.9 and animal-affected sediments 3/19.1
Amplicons from samples with increased intensity of land use were largely actinomycetal. Sixty-seven of 77 clones from cultivated (Colcultivated-Rozo), animal-affected (wormguts-Rozo, hivewaste-Netolice) and urban (cobblestonesBrussels-Boucher, Č.Budéjovice-streetsediment-Česká) DNA samples were from the Actinomycetes. Lineage-specific analysis with UniFrac confirmed that there was selection for the Actinomycetes in non-soil sediments (P = 0.0002, Fig. 2).
ARDRA Patterns Generated Using the 16S rDNA Single Actinobacterial-Specific Primer System (F-Act/R-Bact)
Having gained information into the actinobacterial community composition and phylogenetic relationships from sequencing, fingerprinting analysis of the 16S rDNA sequences was carried out to enable comparison of the many sample sites. TaqI ARDRA was used as the actinobacterial and verrucomicrobial amplicons possessing markedly different banding patterns. Simulated TaqI restriction of 65 actinobacterial sequences found that for all but 12 of the sequences, there were no bands between 94 and 320 bp. Six of the sequences with bands in this range were non-actinomycetal Actinobacteria and four of these were from the SOSED group. Simulated TaqI restriction of all 71 verrucomicrobial clones and their closest matches gave bands in the 95- 320-bp range (e.g. 120, 131 and 181 bp; Supplemental data 1 and 3).
The ARDRA patterns of the different soils and sediments that were investigated in this study clustered into three groups. One group contained cultivated soils, street sediments and animal sediments and possessed few bands in the 94- 320-bp range. A second group contained most of the uncultivated soils and possessed many bands in the 94-320-bp range. Finally, an intermediate group containing animal-affected sediments, soils and street sediments was also identified (Fig. 3). These ARDRA patterns confirmed the F-Act/R-Bact cloning and sequencing results, demonstrating that verrucomicrobial amplicons were more common in soil PCR products and that actinobacterial amplicons were more common in animal-affected and street sediment PCR products.
UPGMA tree of amplified ribosomal DNA restriction analysis (ARDRA) patterns generated using the F-Act/R-Bact primers of Heuer et al. [15] and TaqI digestion. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). For sample descriptions, see Tables 1 and 2
T-RLFP Patterns Generated Using the 16S rDNA Double Actinobacterial (F-Act/R-Act) Primers
Both forward and reverse T-RFLP patterns were used to characterise the F-Act/R-Act 16S rDNA amplicons. Reverse T-RFLP patterns showed the distribution of the SOSED group, whilst forward T-RFLP patterns gave a picture of the actinobacterial community structure.
Reverse T-RFLP
Simulated HhaI restriction of all 84 actinomycetal clones and their BLAST matches showed reverse T-RFLP peaks between 348 and 385 bases (ESM Supplemental data 4 and 5), with all but three peaks between 374 and 385 bases (Colforest/garden-CIATbiotech8, 352 bases; Netolice-hivewaste8, 348 bases; and Micromonospora aurantiaca, 349 bases). Reverse T-RFLP peaks of 374–385 bases were also found among the non-actinomycetal actinobacterial clones, notably the five Microthrix-related clones. However, most non-actinomycetal actinobacterial clones had reverse T-RFLP peaks outside the 374- 385-base range. The most common reverse SOSED group T-RFLP peak was at 186–190 bases (42 of 60 clones), with other peaks at 148, 239–242 and 376–390 bases. For reverse T-RFLP, all peaks outside the 348- 390-base range thus represent non-actinomycetal Actinobacteria. These peaks are mainly representative of the SOSED group.
All but one of the reverse T-RFLP patterns clustered in three groups, two from soil and the third from street and animal-affected sediments. A single sample of environmental DNA from vermicompost (Czech-vermicompost) gave two bands at 262 and 272 bp, which were unrelated to any environmental clones (Fig. 4).
UPGMA tree of terminal restriction fragment polymorphism (T-RFLP) patterns generated using the A3R primer of Monciardini et al. [30] F-Act/R-Act primers and HhaI digestion. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). For sample descriptions, see Tables 1 and 2
Forward T-RFLP
Identical forward T-RFLP HhaI peaks were often produced by different phylogenetic groups; however, patterns clustered clearly by land use.
Simulated HhaI restriction of BLAST matches to the SOSED group and of Norcardioidaceae sequences from the ribosomal database programme both gave forward T-RFLP fragments of 147 bases. A Siberian OS clone gave a peak at 145 bases.
Forward T-RFLP patterns clustered by land use with an uncultivated soil cluster, cultivated soil cluster and cluster of street and animal-affected sediments. Worm cast T-RFLP patterns grouped between cultivated and forested soils; the microcosm that they were from contained earthworms from Colforest-Rozo1 and 2, and soil from Colcultivated-Rozo2. A single street sediment (Brussels-cobblestones-Beurs) clustered with cultivated soils; however, most T-RFLP patterns from street and animal-affected sediments not only clustered away from soils but also differed more from each other (30–60% similarity) than soil patterns (65–90% similarity). Several patterns (e.g. Czech-Vermicompost) did not cluster with any of the others (Fig. 5).
UPGMA tree of terminal restriction fragment polymorphism (T-RFLP) patterns generated using the F-243 primer of Monciardini et al’s [30] F-Act/R-Act primers and HhaI digestion. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). Two sequenced clones are also included for comparison. For sample descriptions, see Tables 1 and 2
Canonical Correspondence Analysis of Fingerprints
As clustering suggested that land use controlled ARDRA and T-RFLP patterns, this was tested using a constrained analysis (RDA) with pH, log % clay and log % organic matter as supplementary variables to interpret their effect as well as with a Monte Carlo permutation test to test the significance of land use. Tests were performed on samples for which pH, organic matter and clay content had been determined (i.e. Netolice-Hivewaste, all urban sediments and soils) as well as soils alone (Table 3).
RDA analysis showed that land use was correlated with ARDRA and forward and reverse T-RFLP patterns and that clay content in turn was closely correlated with land use (Fig. 6a–c), with land use explaining 30%, 48% and 55% of variance (in each case, P values were 0.002). As street sediments were unusually sandy and alkaline, an RDA analyses of only soils was carried out. Again, land use controlled T-RFLP patterns, explaining 28%, 37% and 18% of variance with P values of 0.008, 0.002 and 0.004, respectively. However, there was poor correlation to log clay content and a much stronger correlation to log organic matter (Fig. 6d–f).
Redundancy analysis ordination plots of fingerprint patterns for all characterised samples. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). a Amplified ribosomal DNA restriction analysis (ARDRA) patterns for all characterised samples. b Reverse terminal restriction fragment polymorphism (T-RFLP) patterns for all characterised samples. c Forward terminal restriction fragment polymorphism (T-RFLP) patterns for all characterised samples. d Amplified ribosomal DNA restriction analysis (ARDRA) patterns for all soils. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green). e Reverse terminal restriction fragment polymorphism (T-RFLP) patterns for all soils. f Forward terminal restriction fragment polymorphism (T-RFLP) patterns for all soils
Land use thus appeared to control the actinobacterial ARDRA and T-RFLP patterns. However, either the controlling factors varied across the gradient of land use or there was a controlling factor that was not measured.
Sequences Generated with the Eubacterial Primers of Marchesi et al. [28]
To determine if changes in the actinobacterial community structure found with actinobacterial-specific primers are accompanied by an increase in the actinobacterial population, two environments soil and street sediments were more intensively characterised with eubacterial primers. Whilst Actinobacteria were more common in street sediments than soil, the bacterial communities of street sediments varied considerably.
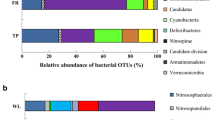
Four hundred seventy-one Eubacterial amplicons from six soils (three cultivated/three forest) and six street sediments (three street dust/three sediments in place) were analysed. Eighty-seven per cent of sequences were from seven orders: the Actinobacteria (126), Alpha proteobacteria (89), Acidobacteria (75), Gamma proteobacteria (59), Bacterioidetes (28), Firmicutes (20) and Delta proteobacteria (15). Whilst UniFrac lineage analysis confirmed that the Actinobacteria were specific to street sediments (P < 0.0000) and that the Acidobacteria were specific to soils (P < 0.0000, Table 4), the gamma and delta proteobacterial clusters interleaved with each other, making lineage-specific analysis impossible. In two samples (České.Budějovice-streetdust-Koh-i-Noor, Streetsediment-ClockFaisalabad), Actinomycetes made up 95–100% of clones sequenced. When these two samples were removed from UniFrac lineage analysis, the Actinobacteria were still specific to street sediments (P < 0.002). Sixty-four per cent (81 out of 127) of actinobacterial clones were from the Micrococcinae (Fig. 7), whereas only 2.7% and 0.64% of clones generated with the F-Act/R-Bact and F-Act/R-Act primers, respectively, were from the Micrococcinae.
Neighbour-joining tree of Actinobacterial 16S clones generated with the F-Act/R-Act primers of Marchesi et al. [28]. Bootstrap values below 50 are not shown. Samples are colour coded as: soils (blue), insect- and earthworm-associated sediments (orange), street sediments (red)
UniFrac Jackknife clustering of the 12 clone libraries found that the three cultivated soils clustered together. The two actinomycetal-rich street dusts clustered together; however, the dust sample from Groningen railway station clustered with the sediment that appeared to be settled from the Budapest railway station. The two railway station communities were dominated by the Bacterioidetes and Gamma proteobacteria (Pseudomonas and in particular the Psychrobacter, ESM Supplemental data 6).The two cobblestone samples differed from each other and all other samples. Brussels-Cobblestones-Boucher contained many Firmicutes (12/42), whilst České.Budějovice-cobblestones-Česka resembled the soil communities (Fig. 8).
Unifrac Jackknife environmental clustering of the 12 eubacterial clone libraries generated with the primers of Marchesi et al. (1998) using the weighted Unifrac algorithm. Jackknife values below 50 are not shown. Samples are colour coded as: forest soils (blue), cultivated soils (sea green), street sediments (red)
Discussion
The results from the three primer sets each tell a different phylogenetic story; however, they give a consistent ecological message. The F-Act/R-Bact primers show a switch from the Verrucomicrobia to the Actinobacteria, increasing from [Uncultivated soils][Cultivated soils][Street sediments/Animal affected sediments]; the F-Act/R-Act primers show a switch from the actinobacterial (but non-actinomycetal) SOSED group to the actinomycetal Norcardioides, increasing from [Uncultivated soils][Cultivated soils][Street sediments/Animal-affected sediments]; and sequences generated with bacterial primers from street sediments were enriched with the actinomycetal Micrococcinae compared to soils.
These results are supported by publications that have looked at a narrower range of soils and land uses. Buckley and Schmidt [4] found that the Verrucomicrobial population of soils decreased with cultivation. Whilst our results show that cultivation changes the actinobacterial community composition, there was no statistically significant evidence that cultivation increases the actinobacterial fraction of the bacterial community. In contrast, Lauber et al. [25] compared four land uses in Georgia, USA—cultivated, pasture, pine plantation and mixed wood forest—characterising the entire bacterial community using Q-PCR and found that the Actinobacteria were more common in pasture and cultivated soils. Waldrop et al. [41] found that an actinomycete-specific PLFA more than doubled on the conversion of Tahitian forest to pineapple plantation, whilst pH decreased slightly. Burke et al. [5] compared the bacterial communities of forest, pasture and sugarcane in Hawaii, Brazil and Ecuador. Actinomycetes were a larger fraction of the microbial community in agricultural than forest soils. Whilst we know of no other studies that characterise the Actinobacteria of street sediments, molecular and cultivation methods have found that Actinomycetes are common in urban environments such as masonry [37], stained glass [36] and wall paintings [14].
The common theme of these results is that the actinobacterial community is controlled and encouraged by human and animal activities: cultivation, urbanisation and digestion. In this study, urbanisation also had a strong selective effect on the Actinobacteria that occurred in different bacterial communities, suggesting that actinobacterial selection is independent of overall bacterial community structure (Fig. 3). What is not clear is whether there is a single controlling factor for all three of these activities which controls actinobacterial community structure or if cultivation, urbanisation and digestion each change the community structure through different mechanisms to develop distinct actinobacterial habitats.
If there is a single factor controlling actinobacterial community structure in these samples, it is not one of the three that we measured: pH, clay content or organic matter. Ordination plots showed that organic matter controlled actinobacterial community structure when fingerprints from soils were compared; pH appeared to control actinobacterial structure when soils and street sediments were compared.
Actinobacterial fingerprints from acidic animal waste samples clustered with street sediments. We can also safely exclude climate and parent material as overall controlling factors as there was no evidence of clustering by sampling area.
Actinobacterial distribution appears to differ from the distribution of the total bacterial community which is controlled by soil properties such as pH [9] and texture [10] and resemble that of the fungi,which is also increases with the conversion of forest to agriculture [5, 41]. It is possible that similarities in role, such as the development of multicellular structures such as filaments/hyphae, mean that there is a common factor controlling actinobacterial and fungal communities. Urbanisation has also been shown to have a profound effect on fungal community structure, with soils from abandoned Russian medieval cities containing similar fungal populations to modern cities [29]. Marfenina et al. also found that urban fungal communities were more beta diverse and ascribed this to the mosaic of environments and substrates found in cities.
The results of this study also have implications for experimental strategies in applying PCR to environmental DNA from soils and sediments.
These results were generated with a relatively low level of sequencing using three sets of primers; clone libraries of at least 400 clones are needed to characterise the soil bacterial community [20]. This is because the object of our sequencing was not to characterise the bacterial communities of samples but either to interpret ARDRA and T-RFLP fingerprinting or to determine if the Actinobacteria were more common in certain samples through UniFrac lineage-specific analysis. It could be argued that the resources spent on fingerprinting would have been better spent on concentrated sequencing to capture more of the rarer fraction (see [1] for both sides of this debate). However, sequences that are rare in clone libraries generated using one primer set may be common in libraries generated with a different primer set [18]. The actinobacterial Micrococcinae were rare in amplicons generated with actinobacterial primers whilst common in our eubacterial amplicons, whereas the Verrucomicrobia, previously found to be common in soil [40], were half of the sequences generated with F-Act/R-Bact primers and absent from sequences generated with eubacterial primers. Relatively shallow sequencing with a range of primers may give a better view of the bacterial community structure than intensive sequencing with a single primer set. Amplification of non-target sequences may yield interesting, if unpredictable, information about the distribution of non-actinobacterial groups. In our case, we found evidence that the AD-3 candidate division is selected for in acid soils, that the Acidobacteria are absent from street sediments (possibly due to alkalinity, they are selected for in acid soils) [21] and that the genus Psychrobacter, known to be common in extreme environments [35], also made up a large fraction of the bacterial community of two railway station samples.
Previous studies of actinobacterial distribution have found that biogeography [42] and pH [26] control community structure in soil, an environment which provides many important ecosystem services. Actinobacterial bioprospecting is to identify environments that are enriched in Actinobacteria, so these studies should include a range of environments which are unimportant for ecosystem services (e.g. in this study, street dust, hive waste). As the results of any PCR reaction are a product of the sample, the DNA extraction method and the primer system used, sampling many contrasting samples including commonly characterised environments such as soil also ensures that one can determine how much results are sample-specific and how much a product of extraction or PCR methods.
References
Bent SJ, Pierson JD, Forney LJ (2007) Measuring species richness based on microbial community fingerprints: the emperor has no clothes. Appl Environ Microbiol 73:2399–2401
Blackwood C, Marsh T, Kim S-H, Paul EA (2003) Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl Environ Microbiol 69:926–932
Bossio DA, Girvan MS, Verchot L, Bullimore J, Borelli T, Albrecht A, Scow KM, Ball AS, Pretty JN, Osborn AM (2005) Soil microbial community response to land use change in an agricultural landscape of Western Kenya. Microb Ecol 49:50–62
Buckley DH, Schmidt TM (2001) Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol Ecol 35:105–112
Burke RA, Molina M, Cox JE, Osher LJ, Piccolo MC (2003) Stable carbon isotope ratio and composition of microbial fatty acids in tropical soils. J Environ Qual 32:198–206
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM (2005) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33:294–296
Ellis RJ, Morgan P, Weightman AJ, Fry JC (2003) Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol 69:3223–3230
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631
Fierer N, Carney KM, Horner-Devine MC, Megonigal JP (2009) The biogeography of ammonia-oxidizing bacterial communities in soil. Microb Ecol 58:435–445
Gelsomino A, Keijzer-Wolters AC, Cacco G, van Elsas JD (1999) Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gel electrophoresis. J Microbiol Methods 38:1–15
Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS (2003) Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol 69:1800–1809
Gomez-Alvarez V, King G, Nuesslein K (2007) Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol Ecol 60:60–73
Graff A, Conrad R (2005) Impact of flooding on soil bacterial communities associated with poplar (Populus sp.) trees. FEMS Microbiol Ecol 53:401–415
Gurtner C, Piñar G, Lubitz W, Swings J, Rölleke S (2000) Comparative analyses of the bacterial diversity on two different biodeteriorated wall paintings by DGGE and 16S rDNA sequence analysis. Int Biodeterior Biodegrad 46:229–239
Heuer H, Krsek M, Baker P, Smalla K, Wellington EM (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63:3233–3241
Hill P, Krištůfek V, Martinez AF, Gallego G (2005) The characterization of wormcasts through of amplified ribosomal DNA (ARDRA) and 16S ribosomal sequencing. In: Tajovský K, Schalghamerský J, Pižl V (eds) Proceedings of the 7th Central European Workshop on Soil Zoology, 14–15 April 2003. Institute of Soil Biology ASCR, České Budějovice, pp 25–29
Holmes AJ, Bowyer J, Holley MP, O'Donoghue M, Montgomery M, Gillings MR (2000) Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol Ecol 33:111–120
Hong SH, Bunge J, Leslin C, Jeon S, Epstein SS (2009) Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 12:1365–1373
İnceoğlu Ö, Hoogwout EF, Hill P, van Elsas JD (2010) Effect of DNA extraction method on the apparent microbial diversity of soil. Appl Environ Microbiol 76:3378–3382
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analysis. ISME J 3:442–453
Kanokratana P, Chanapan S, Pootanakit K, Eurwilaichitr L (2004) Diversity and abundance of bacteria and archaea in the Bor Khlueng Hot Spring in Thailand. J Basic Microbiol 44:430–444
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
LaMontagne MG, Schimel JP, Holden PA (2003) Comparison of subsurface and surface soil bacterial communities in California grassland as assessed by terminal restriction fragment length polymorphisms of PCR-amplified 16S rRNA genes. Microb Ecol 46:216–227
Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land use types. Soil Biol Biochem 40:2407–2415
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Marfenina OE, Ivanova AE, Kislova EE, Sacharov (2008) The mycological properties of medieval culture layers as a form of soil ‘biological memory’ about urbanization. J Soils Sediments 8:340–348
Monciardini P, Sosio M, Cavaletti L, Chiocchini C, Donadio S (2002) New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes. FEMS Microbiol Ecol 43:419–429
Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L (2005) Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol 71:6885–6899
Neufeld JD, Mohn WW (2005) Unexpectedly high bacterial diversity in arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags. Appl Environ Microbiol 71:5710–5718
Nováková A, Elhottová D, Krištůfek V, Lukešová A, Hill P, Kováč L, Mochk A, Lùptáčik P (2005) Feeding sources of invertebrates in Ardovská and Domica cave systems. In: Tajovský K, Schalghamerský J, Pižl V (eds) Proceedings of the 7th Central European Workshop on Soil Zoology, 14–15 April 2003. Institute of Soil Biology ASCR, České Budějovice, pp 107–112
Reed DW, FujitaY DME, Blackwelder DB, Sheridan P, Uchida T, Colwell FS (2002) Microbial communities from methane hydrate-bearing deep marine sediments in a Forearc Basin. Appl Environ Microbiol 68:3759–3770
Rodrigues DF, da Jesus EC, Ayala-del-Rio HL, Pellizari VH, Gilichinsky D, Sepulveda-Torres L, Tiedje JT (2009) Biogeography of two cold-adapted genera: Psychrobacter and Exiguobacterium. ISME J 3:658–665
Rölleke S, Gurtner C, Drewello D, Lubitz W, Weissmann R (2000) Analysis of bacterial communities on historical glass by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. J Microbiol Methods 36:107–114
Schabereiter-Gurtner C, Piñar G, Vybiral D, Lubitz W, Rölleke S (2001) Rubrobacter-related bacteria associated with rosy discolouration of masonry and lime wall paintings. Arch Microbiol 176:347–354
Selenska-Pobell S, Kampf G, Flemming K, Radevaand G, Satchanska G (2001) Bacterial diversity in soil samples from two uranium waste piles as determined by rep-APD, RISA and 16S rDNA retrieval. Antonie Leeuwenhoek 79:149–161
Sangwan P, Chen X, Hugenholtz P, Janssen PH (2004) Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov. of the phylum Verrucomicrobia. Appl Environ Microbiol 70:5875–5881
Sangwan P, Kovacs S, Davis KER, Sait M, Janssen PH (2005) Detection and cultivation of soil Verrucomicrobia. Appl Environ Microbiol 71:8402–8410
Waldrop MP, Balser TC, Firestone MK (2000) Linking microbial community composition to function in a tropical soil. Soil Biol Biochem 32:1837–1846
Wawrik B, Kutliev D, Abdivasievna UA, Kukor JJ, Zylstra GJ, Kerkhof L (2007) Biogeography of actinomycete communities and type II polyketide synthase genes in soil collected in New Jersey and Central Asia. Appl Environ Microbiol 73:2982–2989
Ulrich A, Becker R (2006) Soil parent material is a key determinant of the bacterial community structure in arable soils. FEMS Microbiol Ecol 56:430–443
Zhou J, Beicheng X, Huang H, Treves DS, Hauser LJ, Mural RJ, Palumbo A, Tiedje JM (2003) Bacterial phylogenetic diversity and a novel candidate division of two humid region sandy surface soils. Soil Biol Biochem 3:915–924
Zhou J, Bruns M (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Acknowledgements
We thank Dr. P. Šmilauer, Pablo Hardoim and Dr. J. Salles for assistance with CANOCO analysis, Dr. A. Poulain for useful criticism of the manuscript, Dr. H. Šantrůčková for providing the Siberian soil sample, Ota Rauch for advice on sampling in the Czech Republic and Rashid Nazir for providing the Faisalabad Clock tower sample. We are indebted to the Slovak Caves Administration in Liptovský Mikuláš (Slovakia) for the sampling and research permit and to Daniël van Middelkoop and Sarina de Roos who carried out the bacterial cloning and sequencing as part of a work experience programme for the School for Biomedical Laboratory Techniques. Funding for this work was provided by the NATO Programme Security Through Science ESP.EAP.CLG 981785, the Ministry of Education, Youth and Sports of the Czech Republic (LC06066, 2B06154) and the Research Plan of the Institute of Soil Biology (AV 0Z 6066 0521).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Data 1
Neighbour-joining tree of Verrucomicrobial 16S sequences generated with the F-Act/R-Bact primers of Heuer et al. [15]. Bootstrap values below 50 are not shown. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). Simulated TaqI ARDRA band positions are shown for each sequence (e.g. Czcultivated-netoliceH5 81/120/133/205/278/368). For sample descriptions, see Tables 1 and 2 (GIF 76 kb)
Supplemental Data 3
Neighbour-joining tree of actinobacterial 16S sequences generated with the F-Act/R-Bact primers of Heuer et al. [15]. Bootstrap values below 50 are not shown. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm0associated sediments (orange), street sediments (red). Simulated TaqI amplified ribosomal DNA restriction analysis (ARDRA) band positions are shown for each sequence (e.g. Czpasture-KrumlovH7 81/328/364/392). For sample abbreviations, see Tables 1 and 2 (GIF 134 kb)
Supplemental Data 4
Neighbour-joining tree of non-actinomycetal actinobacterial 16S clones generated with the F-Act/R-Act primers of Monciardini et al. (2003). Bootstrap values below 50 are not shown. Simulated T-RFLP values are shown for all matches where sequence length allows; forward values are in parentheses e.g. (231 bp), reverse values in bold italics, e.g. 375 bp. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). For sample abbreviations, see Tables 1 and 2 (GIF 103 kb)
Supplemental Data 5
Neighbour-joining tree of actinomycetal actinobacterial 16S clones generated with the F-Act/R-Act primers of Monciardini et al. (2003). Bootstrap values below 50 are not shown. Simulated T-RFLP values are shown for all matches were sequence length allows, forward values are in parentheses, e.g. (231 bp), reverse values in bold italics, e.g. 375 bp. Samples are colour coded as: forest/pasture soils (blue), cultivated soils (sea green), insect- and earthworm-associated sediments (orange), street sediments (red). For sample abbreviations, see Tables 1 and 2 (GIF 90 kb)
Supplemental Data 6
Neighbour-joining tree of Psychrobacter 16S clones generated with the eubacterial primers of Marchesi et al. (1998). Bootstrap values below 50 are not shown. For sample abbreviations, see Table 2 (GIF 80 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hill, P., Krištůfek, V., Dijkhuizen, L. et al. Land Use Intensity Controls Actinobacterial Community Structure. Microb Ecol 61, 286–302 (2011). https://doi.org/10.1007/s00248-010-9752-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9752-0