Abstract
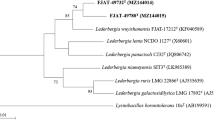
Two Gram-staining positive strains, FJAT-49825T and FJAT-50051T were isolated from a citrus rhizosphere soil sample. Strains FJAT-49825T and FJAT-50051T showed the highest 16S rRNA gene sequence similarity with the type strain of Neobacillus cucumis (98.4–98.5%). The 16S rRNA gene sequence similarity between strains FJAT-49825T and FJAT-50051T was 99.8%. Strain FJAT-49825T optimally grew at 35 °C, pH 6.0 in the absence of NaCl while strain FJAT-50051T grew at 40 °C, pH 7.0 and in presence of 2% NaCl (w/v). Both strains contained meso-2,6-diaminopimelic acid as the cell-wall diamino acid. The respiratory quinone of strains FJAT-49825T and FJAT-50051T was MK-7. The polar lipids of strain FJAT-49825T were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, unidentified aminolipid and unidentified lipid whereas strain FJAT-50051T polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, unidentified phospholipid and unidentified lipid. The major fatty acids (> 10%) in both strains were iso-C15:0 and anteiso-C15:0. The genomic DNA G + C content of strains FJAT-49825T and FJAT-50051T were 40.8 and 41.1%, respectively. The average nucleotide identity and digital DNA–DNA hybridization value between strains FJAT-49825T and FJAT-50051T and with other members of the genus Neobacillus were lower than the cut-off value (95–96/70%) for interspecies identity. Based on the results, strains FJAT-49825T and FJAT-50051T represent two novel species of the genus Neobacillus, for which the names Neobacillus rhizophilus sp. nov. and Neobacillus citreus sp. nov. are proposed. The type strains are FJAT-49825T (= GDMCC 1.2592T = JCM 34834T) and FJAT-50051T (= GDMCC 1.2593T = JCM 34835T).

Similar content being viewed by others
References
Auch AF, von Jan M, Klenk HP, Göker M (2010) Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genom Sci 2(1):117–134. https://doi.org/10.4056/sigs.531120
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. https://doi.org/10.1093/nar/29.12.2607
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2,4-diaminobutyric acid. J Appl Bacteriol 48:459–470. https://doi.org/10.1111/j.1365-2672.1980.tb01036.x
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230. https://doi.org/10.1099/00221287-100-2-221
Dong ZY, Narsing Rao MP, Wang HF, Fang BZ, Liu YH, Li L, Xiao M, Li WJ (2019) Transcriptomic analysis of two endophytes involved in enhancing salt stress ability of Arabidopsis thaliana. Sci Total Environ 686:107–117. https://doi.org/10.1016/j.scitotenv.2019.05.483
Dong ZY, Rao MPN, Liao TJ, Li L, Liu YH, Xiao M, Mohamad OAA, Tian YY, Li WJ (2021) Diversity and function of rhizosphere microorganisms between wild and cultivated medicinal plant Glycyrrhiza uralensis Fisch under different soil conditions. Arch Microbiol 203:3657–3665. https://doi.org/10.1007/s00203-021-02370-y
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO (2015) Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 3:e1319. https://doi.org/10.7717/peerj.1319
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322. https://doi.org/10.2323/jgam.29.319
Kämpfer P, Busse HJ, Glaeser SP, Kloepper JW, Hu CH, McInroy JA (2016) Bacillus cucumis sp. nov. isolated from the rhizosphere of cucumber (Cucumis sativus). Int J Syst Evol Microbiol 66:1039–1044. https://doi.org/10.1099/ijsem.0.000831
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703–704. https://doi.org/10.1038/178703a0
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr 5:2359–2367. https://doi.org/10.1080/01483918208067640
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. https://doi.org/10.1093/nar/gkm160
Lechevalier MP, Lechevalier H (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443. https://doi.org/10.1128/jb.94.4.875-883.1967
Liu B, Liu GH, Hu GH, Chen MC (2014) Bacillus mesonae sp. nov., isolated from the root of Mesona chinensis. Int J Syst Evol Microbiol 64:3346–3352. https://doi.org/10.1099/ijs.0.059485-0
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. https://doi.org/10.1093/nar/25.5.955
Luo R, Liu B, Xie Y et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. https://doi.org/10.1186/2047-217X-1-18
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50(D1):D801–D807. https://doi.org/10.1093/nar/gkab902
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95. https://doi.org/10.1111/j.1365-2672.1979.tb01172.x
Morgan JAW, Bending GD, White PJ (2005) Biological costs and benefits to plant-microbe interactions in the rhizosphere. J Exp Bot 56:1729–1739. https://doi.org/10.1093/jxb/eri205
Murray RGE, Doetsch RN, Robinow CF (1994) Determinative and cytological light microscopy. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 21–41
Narsing Rao MP, Dong ZY, Kan Y, Dong L, Li S, Xiao M, Kang YQ, Zhang K, Li WJ (2020) Description of Paenibacillus tepidiphilus sp. nov., isolated from a tepid spring. Int J Syst Evol Microbiol 70:1977–1981. https://doi.org/10.1099/ijsem.0.004004
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M (2020) List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612. https://doi.org/10.1099/ijsem.0.004332
Patel S, Gupta RS (2020) A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int J Syst Evol Microbiol 70:406–438. https://doi.org/10.1099/ijsem.0.003775
Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK (2016) Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. https://doi.org/10.1039/C5AY02550H
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. Microbial ID Inc, Newark
Tang R, Zhang Q, Narsing Rao MP, Liu GH, Che JM, Lei M, Liu B, Li WJ, Zhou SG (2021) Neobacillus sedimentimangrovi sp. nov., a thermophilic bacterium isolated from mangrove sediment. Curr Microbiol 78:1039–1044. https://doi.org/10.1007/s00284-021-02360-9
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Zhang MY, Cheng J, Cai Y, Zhang TY, Wu YY, Manikprabhu D, Li WJ, Zhang YX (2017) Bacillus notoginsengisoli sp. nov., a novel bacterium isolated from the rhizosphere of Panax notoginseng. Int J Syst Evol Microbiol 67:2581–2585. https://doi.org/10.1099/ijsem.0.001975
Zuo J, Zu M, Liu L, Song X, Yuan Y (2021) Composition and diversity of bacterial communities in the rhizosphere of the Chinese medicinal herb Dendrobium. BMC Plant Biol 21:127. https://doi.org/10.1186/s12870-021-02893-y
Acknowledgements
We are very grateful to Professor Oren for the nomenclature help.
Funding
This work was supported by a grant from Fujian Special Fund for Scientific Research Institutes in the Public Interest (2020R1034001), Fujian Academy of Agricultural Sciences (GJPY2019003) and Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (No. 311021006). WJL was also supported by Introduction project of high-level talents in Xinjiang Uygur Autonomous Region.
Author information
Authors and Affiliations
Contributions
G-HL designed the experiments. G-HL, Q-QC, HS, J-MC performed the chemotaxonomic experiments. MPNR carried out genome analysis, wrote and drafted the manuscript. B-L and W-JL guided the project and finalized the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This article does not contain any studies related to human participants or animals.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, GH., Narsing Rao, M.P., Chen, QQ. et al. Neobacillus rhizophilus sp. nov. and Neobacillus citreus sp. nov., isolated from the citrus rhizosphere soil. Arch Microbiol 204, 281 (2022). https://doi.org/10.1007/s00203-022-02886-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02886-x