Abstract
One hundred and sixty-four accessions representing Czech and Slovak pea (Pisum sativum L.) varieties bred over the last 50 years were evaluated for genetic diversity using morphological, simple sequence repeat (SSR) and retrotransposon-based insertion polymorphism (RBIP) markers. Polymorphic information content (PIC) values of 10 SSR loci and 31 RBIP markers were on average high at 0.89 and 0.73, respectively. The silhouette method after the Ward clustering produced the most probable cluster estimate, identifying nine clusters from molecular data and five to seven clusters from morphological characters. Principal component analysis of nine qualitative and eight quantitative morphological parameters explain over 90 and 93% of total variability, respectively, in the first three axes. Multidimensional scaling of molecular data revealed a continuous structure for the set. To enable integration and evaluation of all data types, a Bayesian method for clustering was applied. Three clusters identified using morphology data, with clear separation of fodder, dry seed and afila types, were resolved by DNA data into 17, 12 and five sub-clusters, respectively. A core collection of 34 samples was derived from the complete collection by BAPS Bayesian analysis. Values for average gene diversity and allelic richness for molecular marker loci and diversity indexes of phenotypic data were found to be similar between the two collections, showing that this is a useful approach for representative core selection.





Similar content being viewed by others
References
Baranger A, Aubrey G, Aran G, Laine AL, Deniot G, Potier J, Burstin J (2004) Genetic diversity within Pisum sativum using protein and PCR-based markers. Theor Appl Genet 108:1309–1321
Beaumont MA, Rannala B (2004) The Bayesian revolution in genetics. Nature Rev 5:251–261
Beckmann JS, Soller M (1990) Toward a unified approach to genetic mapping of eukaryotes based on sequence tagged microsatellite sites. Biotechnology 10:930–932
Bredemeijer GMM, Cooke RJ, Ganal MW, Peeter R, Isakk P, Noordijk Y, Rendell S, Jackson J, Roder MS, Wendehake K, Dijcks M, Amelaine M, Wickaert V, Bertrand L, Vosman B (2002) Construction and testing of a microsatellite database containing more than 500 tomato varieties. Theor Appl Genet 105:1019–1026
Brown AHD, Spillane C (1999) Implementing core collections––principles, procedures, progress, problems and promise. In: Johnson RC, Hodgkin T (eds) Core collections for today and tommorow. IPGRI, Rome
Brown AHD, Weir BS (1983) Measuring genetic variability in plant populations. In: Tanksley SD, Orton TJ (eds) Isozymes in plant genetics and breeding, part A. Elsevier, Amsterdam
Burstin J, Deniot G, Potier J, Weinachter C, Aubert G, Baranger A (2001) Microsatellite polymorphism in Pisum sativum. Plant Breed 120:311–217
Corander J, Martiinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecology 15:2833–2843
Corander J, Waldmann P, Marttinen P, Sillanpää MJ (2004) BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 20:2363–2369
Corander J, Gyllenberg M, Koski T (2007) Random partition models and exchangeability for Bayesian identification of population structure. Bull Math Biol 69:797–815
Donini P, Law JR, Koebner RMD, Reeves JC, Cooke RJ (2000) Temporal trends in the diversity of UK wheat. Theor Appl Genet 100:912–917
Ellis THN, Poyser SJ, Knox MR, Vershinin AV, Ambrose MJ (1998) Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol Gen Genet 260:9–19
Erskine W, Muehlbauer FJ (1991) Allozyme and morpfological variability, outcrossing rate and core collection formation in lentil germplasm. Theor Appl Genet 83:119–125
Esquinas-Alacazar J (2005) Protecting crop genetic diversity for food security: political, ethical and technical challenges. Nature Rev Genet 6:946–953
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Flavell AJ, Knox MR, Pearce SR, Ellis THN (1998) Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J 16:643–650
Ford R, LeRoux K, Itman C, Brouwer JB, Tayler PWJ (2002) Diversity analysis and genotyping in Pisum with sequence tagged microsatellite (STSM) primers. Euphytica 124:397–405
Flavell AJ, Bolshakov VN, Booth A, Jing R, Russell J, Ellis THN, Isaac P (2003) A microarray-based high throughput molecular marker genotyping method: the tagged microarray marker (TAM) approach. Nucleic Acids Res e31:e115
Frankel OH, Brown AHD (1984) Current plant genetic resources––a critical appraisal. In: Genetics: new Frontiers, vol IV. Oxford and IBH Publ. Co., New Delhi
Goudet J (1995) FSTAT (ver. 1.2): a computer program to calculate F-statistics. J Heredity 86:485–486
Hu J, Zhu J, Xu HM (2000) Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theor Appl Genet 101:264–268
Jing RC, Knox MR, Lee JM, Vershinin AV, Ambrose M, Ellis THN, Flavell AJ (2005) Insertional polymorphism and antiquity of PDR1 retrotransposon insertions in Pisum species. Genetics 171:741–752
Jing R, Bolshakov VI, Flavell AJ (2007) The Tagged Microarray Marker (TAM) method for high throughput detection of single nucleotide and indel polymorphisms. Nature Protocols 2:168–177
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Kruskal JB (1964) Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 29:1–27
Le Clerc V, Bazante F, Baril C, Guiard J, Zhang D (2005) Assessing temporal changes in genetic diversity of maize varieties using microsatellite markers. Theor Appl Genet 110:294–302
Le Clerc V, Cadot V, Canadas M, Lallemand J, Guerin D, Boulineau F (2006) Indicators to assess temporal genetic diversity in the French Catalogue: no losses for maize and peas. Theor Appl Genet 113:1197–1209
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–398
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, Lejeune-Henaut I, Burstin J (2005) Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111:1022–1031
Maccaferri M, Sanguineti MC, Noli E, Tuberosa R (2005) Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Mol Breed 15:271–289
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Nei M (1973) Analysis of gene diversity in subdivaded populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nersting LG, Andersen SB, von Bothmer R, Gullord M, Jorgensen RB (2006) Morphological and molecular diversity of Nordic oat through one hundred years of breeding. Euphytica 150:327–337
Pavelková A, Moravec J, Hájek D, Bareš I, Sehnalová J (1986) Descriptor list genus Pisum L. RICP Prague. Genové zdroje 32:46
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Reif JC, Melchinger AE, Frish M (2005) Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci 45:1–7
Reynolds J, Weir BS, Cockerham C (1983) Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767–776
Rohlf F (2006) NTSYSpc: Numerical Taxonomy System (ver. 2.2). Exeter Publishing, Ltd., Setauket
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Roussel V, Leisova L, Exbrayat F, Stehno Z, Balfourier F (2005) SSR allelic diversity changes in 480 European bread wheat varieties released from 1840 to 2000. Theor Appl Genet 111:162–170
Roussel V, Koenig J, Beckert M, Balfourier F (2006) Molecular diversity in French bread wheat accessions related to temporal trends and breeding programmes. Theor Appl Genet 108:920–930
Shannon CE, Weaver W (1962) The mathematical theory of communication. Univer. of Illinois Press, Urbana
Simioniuc D, Uptmoor R, Friedt W, Ordon F (2002) Genetic diversity and relationships among pea cultivars revealed by RAPDs and AFLPs. Plant Breed 121:429–435
Smýkal P (2006) Development of an efficient retrotransposon-based fingerprinting method for rapid pea variety identification. J Appl Genet 47:221–230
Smýkal P, Valledor L, Rodríguez R, Griga M (2007) Assessment of genetic and epigenetic stability in long-term in vitro shoot culture of pea (Pisum sativum L.). Plant Cell Rep 26:1985–1998
SPSS for Windows, Rel. 12.0.1 (2003) SPSS Inc., Chicago
StatSoft Inc (2006) STATISTICA (data analysis software system), version 7.1. http://www.statsoft.com
Tanksley SD, McCouch SR (1997) Seed bank and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tar’an B, Zhang C, Wankertin T, Tullu A, Vandenberg A (2005) Genetic diversity among varietis and wild species accessions of pea (Pisum sativum L.) based on molecular markers, and morphological and physiological characters. Genome 48:257–272
Tohme J, Jones P, Beebe S, Iwanaga M (1995) The combined use of agroecological and characterization data to establish the CIAT Phaseolus vulgaris core collection. In: Hodgkin T, Brown AHD, van Hintum TJL, Morales EAV (eds) Core collections of plant genetic resources. Wiley, Chichester, pp 95–107
Van Hintum TJL (1999) The general methodology for creating a core collection. In: Johnson RC, Hodgkin T (eds) Core collections for today and tomorrow. International Plant Genetic Resources Institute, Rome, pp 10–17
Vavilov NI (1926) Studies on the origin of cultivated plants, Bulletin of Applied Botany, vol 26. Leningrad, USSR
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang JC, Hu J, Xu HM, Zhang S (2007) A strategy on constructing core collections by least distance stepwise sampling. Theor Appl Genet 115:1–8
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BT, Powell W (1997) Genetic distribution of BARE-1 retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253:687–694
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphism amplified by arbitrary primers are usefull as genetic markers. Nucleic Acids Res 18:6531–6535
Wright S (1965) The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution 19:395–420
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction. Genomics 20:176–183
Acknowledgments
This work was financially supported by Ministry of Education of Czech Republic research project MSMT 2678424601 and Czech Ministry of Agriculture project no. 33083/03-3000. Technical support of Mrs. L.Vítámvásová, E. Fialová, M. Vachatová and E. Kamlerová are greatly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. A. Hoisington.
Electronic supplementary material
Below is the link to the electronic supplementary material.
List of
Pisum sativum accessions used in analysis. Accession number, pedigree, date of entry into collection, breeding period, accession type (breeding line, wild accession, land race, variety) and user type (fodder and dry-seed) are indicated. A set of 34 accessions forming the deduced core collection based upon BAPS analysis is indicated (DOC 44 kb)
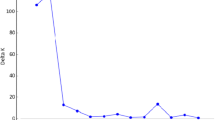
Ward hierarchical classification performed on the combined SSR and RBIP molecular distance matrix.
The silhouette method calculation of the most probable cluster number (9) is indicated in the upper corner (DOC 127 kb)
Class frequency distributions;
A. 15 qualitative morphological characters B. 18 quantitative morphological characters in the full set of 164 accessions (DOC 47 kb)
A.
Eigenvalue matrices and vectors of principal components for; A. 9 qualitative characters. B. 18 quantitative characters, of field and fodder pea assessed in field trials (DOC 107 kb)
Ward hierarchical ascendant classification of morphological characters calculated by simple matching coefficient.
4 clusters revealed by the silhouette method as the most homogeneous solution are indicated (DOC 23 kb)
Rights and permissions
About this article
Cite this article
Smýkal, P., Hýbl, M., Corander, J. et al. Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theor Appl Genet 117, 413–424 (2008). https://doi.org/10.1007/s00122-008-0785-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0785-4