Abstract
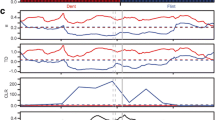
The timing of transition from vegetative growth to flowering is important in nature as well as in agriculture. One of several pathways influencing this transition in plants is the gibberellin (GA) pathway. In maize (Zea mays L.), the Dwarf8 (D8) gene has been identified as an orthologue of the gibberellic acid-insensitive (GAI) gene, a negative regulator of GA response in Arabidopsis. Nine intragenic polymorphisms in D8 have been linked with variation in flowering time of maize. We tested the general applicability of these polymorphisms as functional markers in an independent set of inbred lines. Single nucleotide primer extension (SNuPe) and gel-based indel markers were developed, and a set of 71 elite European inbred lines were phenotyped for flowering time and plant height across four environments. To control for population structure, we genotyped the plant material with 55 simple sequence repeat markers evenly distributed across the genome. When population structure was ignored, six of the nine D8 polymorphisms were significantly associated with flowering time and none with plant height. However, when population structure was taken into consideration, an association with flowering time was only detected in a single environment, whereas an association across environments was identified between a 2-bp indel in the promoter region and plant height. As the number of lines with different haplotypes within subpopulations was a limiting factor in the analysis, D8 alleles would need to be compared in isogenic backgrounds for a reliable estimation of allelic effects.




Similar content being viewed by others
References
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Cardon LR, Palmer LJ (2003) Population stratification and spurious allelic associations. Lancet 361:598–604
Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 locus of barley cv. Himalaya. Molecular and physiological characterization. Plant Physiol 129:181–190
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fehr WR (1987) Principles of cultivar development: theory and technique, 1st edn. Macmillan, New York
Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132:1107–1114
Goodman MM, Stuber CW (1983) Races of maize. VI. Isozyme variation among races of maize in Bolivia. Maydica 28:169–187
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graphic Statist 5:299–314
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13:999–1010
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347
Kimura M, Ohta T (1978) Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc Natl Acad Sci USA 75:2868–2872
Knapp SJ, Bridges WC (1987) Confidence interval estimators for heritability for several mating and experiment designs. Theor Appl Genet 73:759–763
Koch CA, Anderson D, Moran MF, Ellis CE, Pawson T (1991) SH2 and SH3 domains: elements that control interactions of cytoplasmic signalling proteins. Science 252:668–674
Le Corre V, Roux F, Reboud X (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19:1261–1271
Lübberstedt T, Melchinger AE, Fähr S, Klein D, Dally A, Westhoff P (1998) QTL mapping in testcrosses of flint lines of maize. III. Comparison across populations for forage traits. Crop Sci 38:1278–1289
Maurer H-P, Melchinger AE, Frisch M (2004) plabsoft: software for simulation and data analysis in plant breeding. In: Vollmann J, Grausgruber H, Ruckenbauer P (eds) 17th EUCARPIA general congress 2004. BOKU-University of Natural Resources and Applied Life Sciences, pp 359–362
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14[Suppl]:S111–S130
Olsen KM, Halldorsdottir SS, Stinchcombe JR, Weinig C, Schmitt J, Purugganan MD (2004) Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167:1361–1369
Peng J, Harberd NP (1993) Derivative alleles of the Arabidopsis Gibberellin-Insensitive (gai) mutation confer a wild-type phenotype. Plant Cell 5:351–360
Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194–3205
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000a) Association mapping in structured populations. Am J Hum Genet 67:170–181
Pritchard JK, Stephens M, Donnelly P (2000b) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Putterill J, Laurie R, Macknight R (2004) It’s time to flower: the genetic control of flowering time. Bioessays 26:363–373
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Searle SR (1987) Linear models for unbalanced data. Wiley, New York
Silverstone AL, Ciampaglio CN, Sun TP (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the Gibberellin signal transduction pathway. Plant Cell 10:155–170
Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296:285–289
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Utz HF (2003) platstat—a computer program for statistical analysis of plant breeding experiments, version 3A. University of Hohenheim, Stuttgart
Wright S (1978) Evolution and the genetics of populations: variability within and among natural populations, 1st edn. University of Chicago Press, Chicago, pp 91–91
Acknowledgements
We thank the Danish Ministry of Food, Agriculture and Fisheries for Financial support. We also thank E.S. Buckler IV and colleagues for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.C. Becker
Appendices
Appendix 1
The 71 inbred lines included in the analysis with recorded phenotypic dataa at the four locationsb. DMF, DFF and PHT were recorded in EWE and HOH; DMF and DFF were recorded in SÜN, while DFF was recorded in POC
Line name | DMF, EWE | DFF, EWE | PHT, EWE | DMF, HOH | DFF, HOH | PHT, HOH | DMF, SÜN | DFF, SÜN | DFF, POC | |
|---|---|---|---|---|---|---|---|---|---|---|
Flint | F005 | 71.6 | 71.9 | 174.9 | 74.4 | 78 | 187.6 | 72.9 | 77.5 | 74 |
F012 | 65 | 63.9 | 127.9 | 73.1 | 73 | 178.4 | 71 | 71.5 | 74 | |
F013 | 63.9 | 63 | 132.8 | 72.4 | 73 | 182.4 | 70 | 71 | 73 | |
F023 | 74.4 | 75.4 | 134.3 | 77.6 | 81.5 | 199 | 73.1 | 78.5 | 76.5 | |
F030 | 71.7 | 72.1 | 129.6 | 75.9 | 80 | 181.2 | 74 | 76 | 74 | |
F034 | 71.7 | 71.5 | 130.1 | 74.1 | 77.5 | 178.3 | 73.1 | 77.5 | 76 | |
F037 | 69.3 | 68.9 | 148.7 | 73.1 | 74 | 200.2 | 72 | 73.5 | 74.5 | |
F039 | 74.5 | 76.9 | 127.8 | 74.5 | 81.5 | 197.3 | 74 | 80 | 76.5 | |
F040 | 64.1 | 63.8 | 119 | 72 | 71.5 | 165 | 71 | 71.5 | 73 | |
F043 | 71.9 | 71.9 | 125.8 | 74 | 74.5 | 169.7 | 73.7 | 75 | 74.5 | |
F045 | 66.7 | 66.5 | 131.3 | 73 | 75 | 179.2 | 74.5 | 76 | 75.5 | |
F047 | 70.4 | 72.9 | 153.8 | 74.5 | 75 | 177.4 | 73 | 75 | 73 | |
F048 | 71.6 | 71.9 | 142 | 75 | 76.5 | 189.6 | 72.6 | 75.5 | 73 | |
F052 | 67.6 | 68.6 | 141.3 | 74.3 | 77 | 184.1 | 74 | 76.5 | 76.5 | |
F054 | 69.9 | 69.3 | 146.7 | 74.8 | 75 | 190.6 | 72 | 73 | 73.5 | |
Flint/Lancaster | L005 | 78.3 | 78.9 | 127.5 | 77 | 84.5 | 160.2 | 81.5 | 82 | 82 |
L007 | 79.9 | 80.8 | 163.3 | 86.5 | 89.5 | 192.2 | 83 | 82.5 | 85.5 | |
L012 | 78.3 | 81.3 | 191.8 | 82.3 | 87 | 196 | 80 | 84 | 87 | |
L016 | 73.1 | 74.6 | 145.1 | 78 | 81 | 184.6 | 77.5 | 80 | 77.5 | |
L017 | 71.8 | 71.7 | 132 | 75 | 78.5 | 171 | 76.4 | 76.5 | 77 | |
L019 | 73.6 | 74.1 | 145.9 | 78.4 | 80.5 | 180.9 | 79 | 79 | 76 | |
L023 | 73.6 | 74.5 | 161.7 | 79 | 80.5 | 201.9 | 77.7 | 77.9 | 75 | |
L024 | 73.9 | 74.7 | 150.5 | 78.2 | 80 | 182.9 | 75.5 | 78 | 77 | |
L025 | 72.4 | 72.8 | 166.9 | 78.5 | 80 | 187.6 | 77 | 77 | 76.5 | |
L032 | 75.1 | 74.9 | 148.7 | 80 | 82.5 | 181.3 | 77.5 | 79.5 | 77 | |
L035 | 71 | 70.2 | 144.8 | 76 | 79 | 184.8 | 75 | 76 | 76.5 | |
L037 | 73.6 | 73.1 | 151.4 | 77 | 80 | 177.4 | 75.9 | 79 | 77 | |
L041 | 67 | 67.1 | 137.2 | 74.5 | 79 | 175.8 | 72.9 | 75 | 74.5 | |
L043 | 73.6 | 74 | 145 | 78 | 80.5 | 179.2 | 77.5 | 79 | 75 | |
L045 | 73.7 | 76.7 | 159.6 | 81 | 84.5 | 196.1 | 78 | 79.5 | 80.5 | |
L046 | 69.4 | 69.4 | 138 | 75 | 79 | 183.7 | 73.5 | 77 | 73.5 | |
L047 | 66.8 | 65.4 | 146.1 | 73.9 | 75 | 172.4 | 71.5 | 73 | 74.5 | |
L050 | 74.3 | 74 | 168.4 | 82.4 | 83 | 180.1 | 79.9 | 79.5 | 77 | |
Iodent and Iodent/Stiff stalk | P001 | 79.3 | 79.8 | 158 | 83.4 | 85.5 | 185.1 | 80.9 | 85 | 86.5 |
P006 | 73.9 | 75.3 | 157 | 79.4 | 79.5 | 193.6 | 75 | 78.5 | 78.5 | |
P024 | 78.8 | 79.9 | 167.3 | 82.1 | 85 | 187 | 79.5 | 82 | 80.5 | |
P027 | 77.9 | 78.7 | 154.5 | 82.2 | 82.5 | 188.3 | 79.6 | 80.5 | 78 | |
P033 | 77.1 | 76.8 | 149.3 | 80.5 | 82 | 163.9 | 79 | 79.5 | 78 | |
P034 | 76.7 | 76.7 | 163.3 | 78.5 | 80 | 215.5 | 77 | 80.5 | 77.5 | |
P036 | 74.7 | 72.9 | 158 | 79 | 78.5 | 192 | 78.5 | 76.5 | 76 | |
P038 | 73.9 | 75.5 | 121.6 | 80.1 | 83 | 164.6 | 75.1 | 79 | 77 | |
P040 | 77.8 | 78.4 | 148.7 | 82.1 | 86.5 | 182.7 | 78.9 | 80.5 | 80.5 | |
P042 | 77.3 | 76.7 | 160.6 | 80.9 | 81 | 193.7 | 80.5 | 80 | 76.5 | |
P045 | 70.7 | 70.2 | 138.4 | 77.4 | 79 | 179 | 73 | 76 | 75.5 | |
P046 | 69.4 | 68.6 | 139 | 75.5 | 78 | 173.7 | 73 | 77 | 75 | |
P047 | 73.5 | 72.7 | 140.5 | 78.6 | 78.5 | 177.3 | 74.9 | 75.5 | 76.5 | |
P048 | 75.1 | 75.3 | 143.1 | 80.5 | 81.5 | 168.2 | 76.5 | 78 | 81.5 | |
P053 | 75.3 | 77.5 | 144.4 | 80.2 | 83.5 | 166.4 | 76.5 | 80 | 80.5 | |
P060 | 77.4 | 77.3 | 132 | 82.5 | 86 | 187.3 | 79 | 81.5 | 84 | |
P063 | 78.2 | 80.9 | 149 | 83 | 86.5 | 183.7 | 81.4 | 83 | 82 | |
P064 | 79.7 | 80.7 | 160.4 | 83.5 | 86.5 | 190.8 | 79.3 | 83.9 | 86 | |
P065 | 81.1 | 81 | 167.9 | 84.7 | 85 | 204.5 | 80.9 | 82.5 | 86 | |
P066 | 74.7 | 75.7 | 144.2 | 80 | 81 | 189.3 | 79 | 79.5 | 77.5 | |
Stiff stalk | S002 | 74.6 | 74.8 | 142.9 | 81 | 83 | 163.7 | 77 | 78.5 | 76.5 |
S015 | 80 | 80.3 | 148.4 | 84 | 86 | 184.8 | 81.5 | 82.5 | 83.5 | |
S016 | 75.1 | 76.3 | 132.5 | 82.5 | 83.5 | 169.6 | 80 | 81 | 82 | |
S018 | 82.8 | 84 | 155.1 | 89.5 | 90 | 187.2 | 86 | 86.5 | 86.5 | |
S020 | 85 | 85.8 | 168.2 | 90.5 | 90.5 | 187.9 | 86 | 86.5 | 87.5 | |
S028 | 76.5 | 76.5 | 161.1 | 82.5 | 84 | 180.7 | 79.9 | 81 | 80 | |
S033 | 79.7 | 78.9 | 160.3 | 82.9 | 86.5 | 183.1 | 81.5 | 84.5 | 82.5 | |
S035 | 73.9 | 74.3 | 136.5 | 79 | 81 | 169.9 | 77 | 80 | 77 | |
S036 | 73.3 | 75.3 | 144.3 | 80.5 | 83.5 | 166.6 | 78.5 | 80 | 76.5 | |
S040 | 73.4 | 71.8 | 144.4 | 77.5 | 78.5 | 185.9 | 74.5 | 74 | 73 | |
S044 | 79.4 | 79.1 | 197.4 | 82.9 | 83.5 | 206 | 81.1 | 81.5 | 82 | |
S046 | 75 | 74.6 | 138 | 81 | 81.5 | 161.4 | 79.4 | 80 | 78.5 | |
S049 | 76 | 75.1 | 149.6 | 81.5 | 81.5 | 175 | 79.9 | 80 | 80.8 | |
S050 | 78.3 | 78.9 | 166.6 | 81.5 | 83.5 | 180.2 | 75.5 | 80.5 | 78 | |
S058 | 75.1 | 74.7 | 113.3 | 80 | 80.5 | 151.8 | 76.5 | 78 | 74.5 | |
S065 | 72.5 | 72.7 | 132.2 | 77 | 78.5 | 163.8 | 77.1 | 77.5 | 77 | |
S066 | 73 | 73.2 | 155.4 | 75.4 | 79 | 163.7 | 73 | 77 | 76 | |
S067 | 73.7 | 73.3 | 127.1 | 79 | 80 | 162.5 | 79 | 79.5 | 76.5 |
Appendix 2
The SSR analysis was performed with 55 publicly available SSR markers providing an even coverage of the maize genome
Chromosome | Bin | Marker name |
|---|---|---|
1 | 1 | phi427913 |
3 | phi109275 | |
4 | umc1169 | |
6 | umc1122 | |
9 | phi011 | |
11 | phi064 | |
2 | 1 | phi96100 |
3 | umc1555 | |
4 | phi083 | |
8 | phi127 | |
10 | phi101049 | |
3 | 1 | phi104127 |
2 | phi374118 | |
5 | phi053 | |
6 | phi102228 | |
7 | umc1489 | |
9 | umc1641 | |
4 | 1 | phi072 |
1 | phi213984 | |
4 | phi308090 | |
5 | phi079 | |
8 | phi093 | |
10 | umc1180 | |
5 | 2 | phi396160 |
4 | phi331888 | |
5 | phi333597 | |
7 | phi128 | |
9 | umc1153 | |
6 | 0 | umc1143 |
1 | phi423796 | |
3 | umc1887 | |
4 | phi031 | |
7 | phi123 | |
8 | phi089 | |
7 | 0 | umc1545 |
3 | phi114 | |
4 | phi328175 | |
5 | phi069 | |
6 | phi116 | |
8 | 0 | phi420701 |
2 | umc1304 | |
3 | phi121 | |
3 | phi100175 | |
8 | phi015 | |
9 | phi233376 | |
9 | 0 | umc1279 |
3 | phi065 | |
4 | phi032 | |
5 | phi108411 | |
7 | umc1675 | |
10 | 0 | phi041 |
1 | umc1152 | |
3 | phi050 | |
4 | phi084 | |
6 | umc1061 |
Appendix 3
Correspondence between D8 haplotypes (Fig. 2) and the 71 maize lines included in the analysis
Haplotype | Lines |
|---|---|
1 | F012, F013, F034, F045, F048, L016, L035 |
2 | P036, P060, S002, S015, S028, S033, S035, S036, S040, S050 |
3 | P034 |
4 | F040, L024 |
5 | L037 |
6 | L012, L045, L047, L050 |
7 | F005, F023, F030, F037, F039, F043, F047, F052, F054, L005, L007, L017, L019, L023, L025, L032, L041, L043, L046, P001, P006, P024, P027, P033, P038, P040, P042, P045, P046, P047, P048, P053, P064, P065, P066, S016, S018, S020, S044, S046, S058, S065 |
8 | P063, S049, S066, S067 |
Rights and permissions
About this article
Cite this article
Andersen, J.R., Schrag, T., Melchinger, A.E. et al. Validation of Dwarf8 polymorphisms associated with flowering time in elite European inbred lines of maize (Zea mays L.). Theor Appl Genet 111, 206–217 (2005). https://doi.org/10.1007/s00122-005-1996-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-1996-6