Abstract
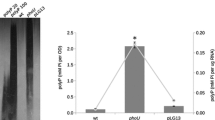
Mutants of Pseudomonas aeruginosa deficient in the utilization of l-proline as the only carbon and nitrogen source have been found to be defective either in proline dehydrogenase activity or in both proline dehydrogenase and Δ1-pyrroline-5-carboxylate dehydrogenase activities of the bifunctional proline degradative enzyme. The latter type of mutants was unable to utilize l-ornithine, indicating that a single Δ1-pyrroline-5-carboxylate dehydrogenase activity is involved in the degradation of ornithine and proline. Proline dehydrogenase and Δ1-pyrroline-5-carboxylate dehydrogenase activities were strongly and coordinately induced by proline. It was excluded that Δ1-pyrroline-5-carboxylate acted as an inducer of the bifunctional enzyme and it was shown that the low level induction observed during growth on ornithine was due to the intracellular formation of proline. The formation of the proline degradative enzyme was shown to be subject to catabolite repression by citrate and nitrogen control.
Similar content being viewed by others
Abbreviations
- EMS:
-
Ethylmethane sulfonate
- NG:
-
N-methyl-N′-nitro-N′-nitrosoguanidine
- P:
-
Minimal medium P
- Pro-DH:
-
Proline dehydro-genase
- P5C:
-
Δ1-Pyrroline-5-carboxylate
- P5C-DH:
-
Δ1-Pyrroline-5-carboxylate dehydrogenase
References
Adelberg EA, Mandel M, Cheng GCC (1965) Optimal conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem Biophys Res Commun 18:788–795
Arst HN, Jones SA, Bailey CR (1981) A method for the selection of deletion mutations in the l-proline catabolism gene cluster of Aspergillus nidulans. Genet Res 38:171–195
Bachmann BJ, Low KB (1980) Linkage map of Escherichia coli K12, edition 6. Microbiol Rev 44:1–56
Brandriss MC, Magasanik B (1980) Proline: an essential intermediate in arginine degradation in Saccharomyces cerevisiae. J Bacteriol 143:1403–1410
Clarke PH, Ornston LN (1975) Metabolic pathways and regulation I. In: PH Clarke, MH Richmond (eds) Genetics and biochemistry of Pseudomonas. Wiley, London, pp 191–261
Day M, Potts JR, Clarke PH (1975) Location of genes for the utilization of acetamide, histidine and proline on the chromosome of Pseudomonas aeruginosa. Genet Res 25:71–78
De Hauwer G, Lavalle R, Wiame JM (1964) Pyrroline dehydrogenase and catabolism regulation of arginine and proline in Bacillus subtilis. Biochim Biophys Acta 81:257–269
Haas, D, Holloway BW (1978) Chromosome mobilization by the R plasmid R68.45: a tool in Pseudomonas genetics. Mol Gen Genet 158:229–237
Haas D, Watson J, Krieg R, Leisinger T (1981) Isolation of an Hfr donor of Pseudomonas aeruginosa PAO by insertion of the plasmid RP1 into the tryptophan synthase gene. Mol Gen Genet 182: 240–244
Holloway BW (1955) Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581
Janssen DB, Herst PM, Joosten HMLJ, van der Drift C (1981) Nitrogen control in Pseudomonas aeruginosa: a role for glutamine in the regulation of the synthesis of NADP-dependent glutamate dehydrogenase, urease and histidase. Arch Microbiol 128:398–402
Krishna RV, Beilstein P, Leisinger T (1979) Biosynthesis of proline in Pseudomonas aeruginosa. Properties of γ-glutamyl-phosphate reductase and 1-pyrroline-5-carboxylate reductase. Biochem J 181:223–230
Krishna RV, Leisinger T (1979) Biosynthesis of proline in Pseudomonas aeruginosa. Partial purification and characterization of γ-glutamyl kinase. Biochem J 181:215–222
Leisinger T, Haas D, Hegarty MP (1972) Indospicine as an arginine antagonist in Escherichia coli and Pseudomonas aeruginosa. Biochim Biophys Acta 262:214–219
Lessie TG, Neidhardt FC (1976) Formation and operation of the histidine-degrading pathway in Pseudomonas aeruginosa. J Bacteriol 93:1800–1810
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Meile L (1982) Prolinkatabolismus in Pseudomonas aeruginosa. Eigenschaften gereinigter Prolin-Dehydrogenase/1-Pyrrolin-5-Carboxylat-Dehydrogenase und Regulation ihrer Synthese. Dissertation Eidgenössische Technische Hochschule Zürich
Menzel R, Roth J (1981a) Purification of the putA gene product. A bifunctional membrane-bound protein from Salmonella typhimurium responsible for the two-step oxidation of proline to glutamate. J Biol Chem 256:9755–9761
Menzel R, Roth J (1981b) Enzymatic properties of the purified putA protein from Salmonella typhimurium. J Biol Chem 256:9762–9766
Menzel R, Roth J (1981c) Regulation of the genes for proline utilization in Salmonella typhimurium: autogenous repression by the putA gene product. J Mol Biol 148:21–44
Mercenier A, Simon J-P, Haas D, Stalon V (1980) Catabolism of l-arginine by Pseudomonas aeruginosa. J Gen Microbiol 116:381–389
Pirt SJ (1975) Principles of microbe and cell cultivation. Blackwell, Oxford
Potts JR, Clarke PH (1976) The effect of nitrogen limitation on catabolite repression of amidase, histidase and urocanase in Pseudomonas aeruginosa. J Gen Microbiol 93:377–387
Prival MJ, Magasanik B (1971) Resistance to catabolite repression of histidase and proline oxidase during nitrogen-limited growth of Klebsiella aerogenes. J Biol Chem 246:6288–6296
Royle P, Matsumoto H, Holloway BW (1981) Genetic circularity of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol 145:145–155
Stanisich V, Holloway BW (1969) Conjugation in Pseudomonas aeruginosa. Genetics 61:327–339
Stanisich V, Holloway BW (1972) A mutant sex factor of Pseudomonas aeruginosa. Genet Res 19:91–108
Tyler B (1978) Regulation of the assimilation of nitrogen compounds. Ann Rev Biochem 47:1127–1162
Voellmy R, Leisinger T (1975) Dual role for N 2-acetylornithine 5-aminotransferase from Pseudomonas aeruginosa in arginine biosynthesis and arginine catabolism. J Bacteriol 122:799–809
Voellmy R, Leisinger T (1976) Role of 4-aminobutyrate aminotrans-ferase in the arginine metabolism of Pseudomonas aeruginosa. J Bacteriol 128:722–729
Watson JM, Holloway BW (1976) Suppressor mutations in Pseudomonas aeruginosa. J Bacteriol 125:780–786
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Meile, L., Soldati, L. & Leisinger, T. Regulation of proline catabolism in Pseudomonas aeruginosa PAO. Arch. Microbiol. 132, 189–193 (1982). https://doi.org/10.1007/BF00508729
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00508729